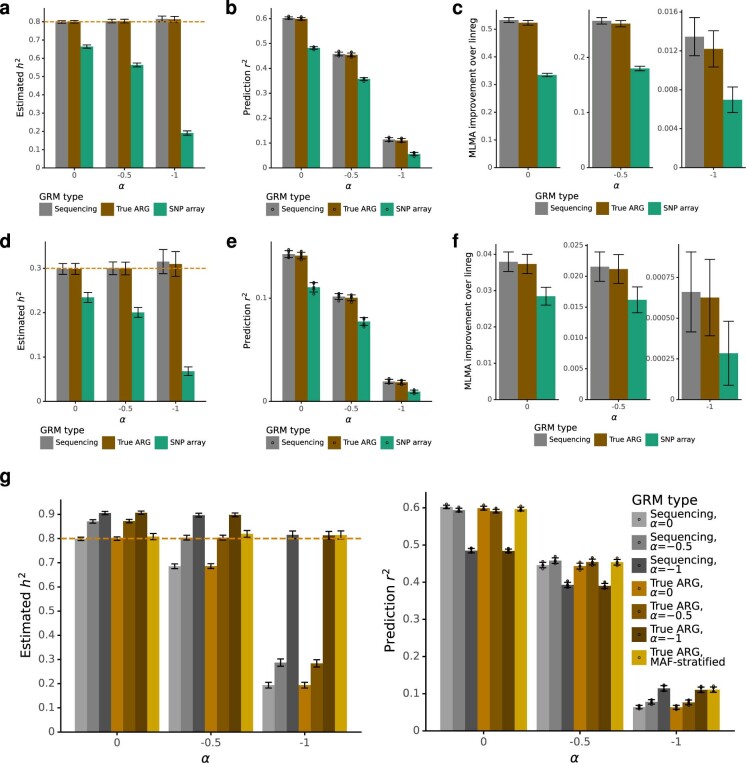
Extended Data Fig. 8. Additional simulations for ground-truth ARG-GRMs.
a-f. As in Fig. 3c, with N = 10,000 haploid samples, except we vary h2 ∈ {0.8, 0.3} and α ∈ {0, −0.5, −1}. a, d. Heritability estimation for a 50 Mb region for h2 = 0.8 (a) and h2 = 0.3 (d). b, e. Polygenic prediction for a 50 Mb region for h2 = 0.8 (b) and h2 = 0.3 (e). c, f. Mixed-model association for 22 chromosomes of 2.5 Mb each for h2 = 0.8 (c) and h2 = 0.3 (f). g. Panels a-f assumed it is possible to infer α and used the true α when building genotype-based or ARG-GRMs. If this value of α is misspecified, heritability estimation is biased and prediction r2 is hampered. This is true both for ARG-GRMs and sequencing GRMs. However, using MAF-stratified ARG-GRMs provides a robust way to estimate the true heritability when α is unknown, and achieves prediction r2 comparable to using the true α (N = 10,000 haploid samples, 50 Mb, h2 = 0.8). For all panels, heritability and prediction experiments involve 5 simulations per bar, and most association experiments involve 50 simulations per bar, except for the h2 = 0.3, α = −1 condition in f, which involved 500 simulations. Data are presented as estimates ± 2 s.e.m., where the estimates are from meta-analysis in the case of heritability estimation and represent means otherwise. Prediction r2 for individual simulations is shown in b and e.