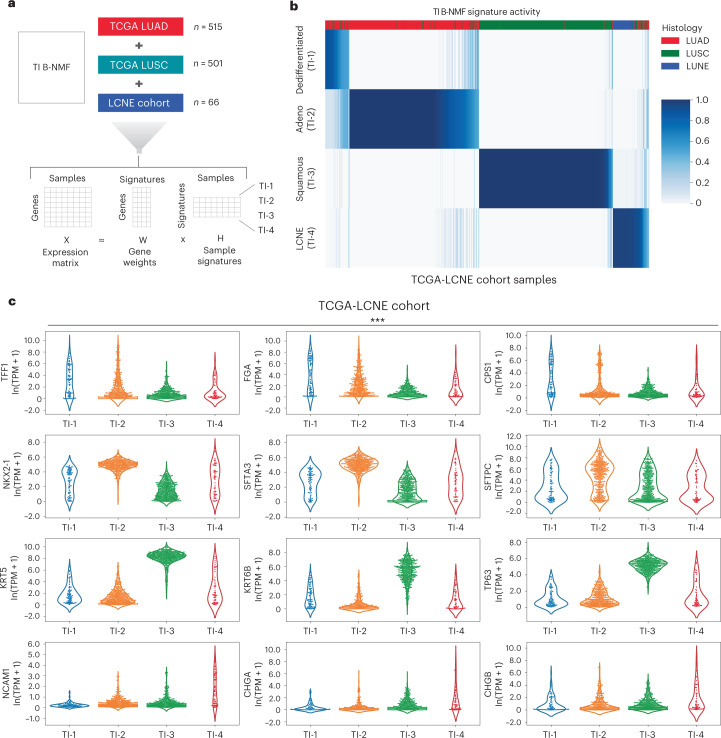
Fig. 4. Derivation of TI NSCLC transcriptional subtypes.
a, Overview of B-NMF approach to the generation of TI subtype signatures. A total of 1,082 RNA-seq samples spanning the three dominant NSCLC histologies were used as input for signature identification. Specifically, the TCGA LUAD and LUSC cohorts were used in addition to a published LCNE Cohort by George et al.63 to generate the combined TCGA-LCNE cohort. b, H-matrix of TCGA-LCNE samples and normalized TI signature activity. c, Violin plots of cancer subtype immunohistochemistry markers based on membership in TI clusters TI-1 (n = 81 samples), TI-2 (n = 433 samples), TI-3 (n = 447) and TI-4 (n = 55). Dedifferentiated (TI-1) samples expressed lower levels of canonical adenocarcinoma and squamous markers, but notably high levels of markers associated with neighboring endodermal lineages (top row). Significance was assessed by the Kruskal–Wallis test (***P < 0.001).