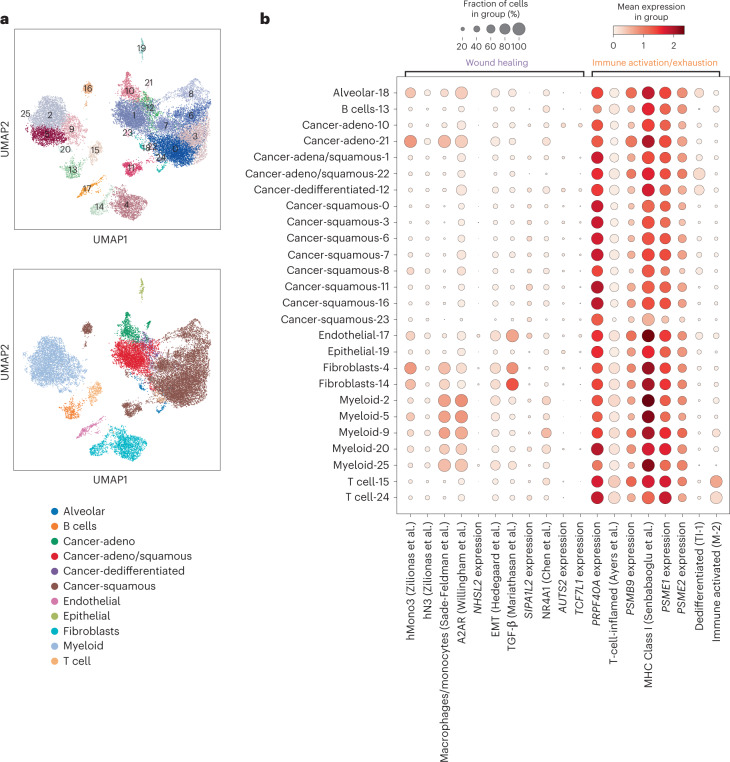
Fig. 7. Exploration of top SU2C-MARK transcriptomic features in single-cell data.
a, Leiden clustering of single-cell RNA-seq data from NSCLC51 colored by cluster ID (upper) or cell-type label (lower). Exploration of tumor markers within the cancer-specific clusters enabled further resolution into NSCLC subtypes, including recapitulation of the dedifferentiated TI-1 subtype identified earlier from bulk RNA-seq data (Cluster 12; Extended Data Fig. 8a). b, Association between cell types identified in NSCLC single-cell data and selected genes and metagenes from the wound healing (C1) and immune activation/exhaustion (C2) feature clusters in the SU2C-MARK cohort or with previously described relationships to immunotherapy response35,45–50. Features within larger correlation blocks in bulk RNA-seq data did not always arise from the same single-cell sources (for example, TGF-β versus macrophages/monocytes in the wound healing cluster, and dedifferentiated TI-1 versus immune-activated M-2 in the immune activation/exhaustion cluster).