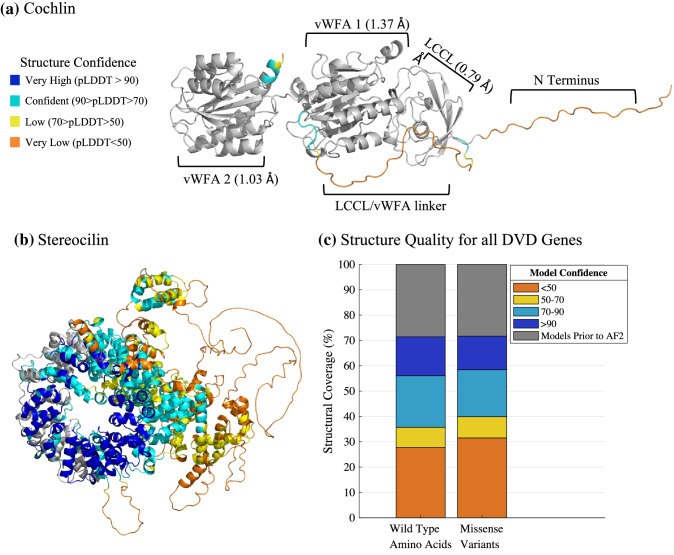
Fig. 1.
Structures and quality of proteins implicated in deafness. AlphaFold2’s novel predicted protein regions are color coded by confidence in the prediction. Gray domains represent homology or experimental structures curated in prior work for a cochlin and b stereocilin. a The root-mean-square deviation (RMSD) of the LCCL and vWFA domains of cochlin (COCH) from AlphaFold2’s domain predictions to the previous models are shown in parentheses. b AlphaFold2 increased protein structural coverage of stereocilin (STRC) from 12 to 100%. However, our work improved the quality of sterocilin’s structure through optimization (described in Methods). As a result of our optimization, the MolProbity score (described in Materials and methods) of the STRC structure improved from 3.07 to 0.98. c Structural model coverage of wild-type amino acids and missense variants for the entire deafness proteome shows that this work increased coverage from < 30% (gray, prior work) to 100% coverage. The stacked bars are color coded based on confidence in the protein structure. The wild-type amino acids and missense variants in the deafness proteome are present in similar proportions across all structural confidence ranges, indicating that specific confidence regions are not enriched for the presence of missense variants