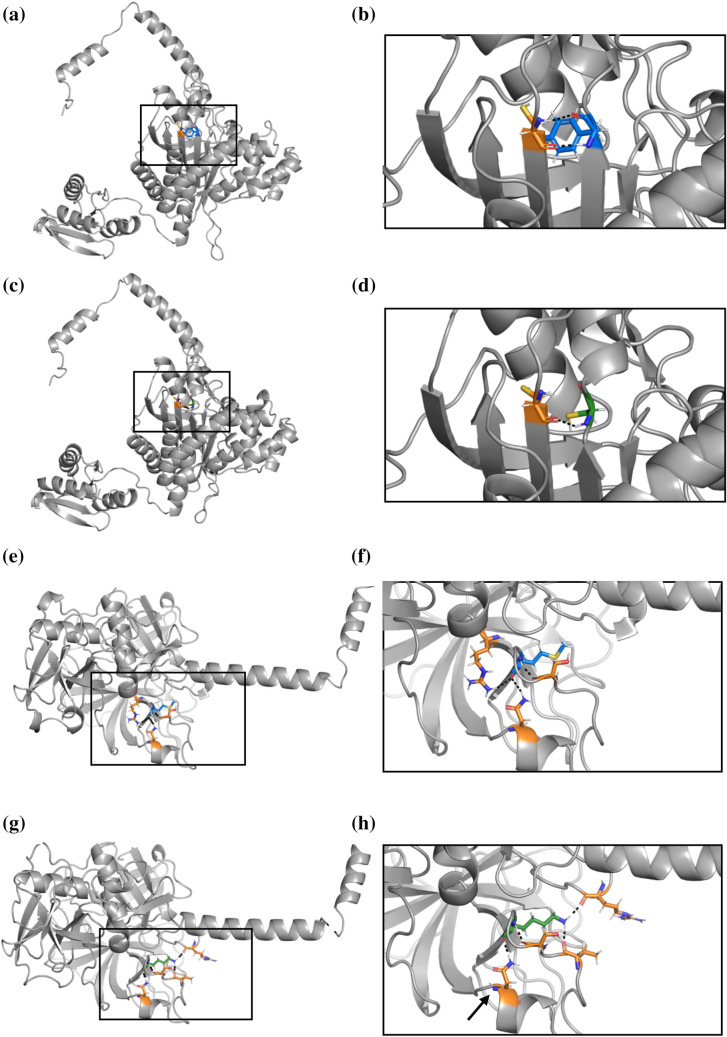
Fig. 6.
The protein structure of HARS2 variant NP_036340.1:p.Tyr364Cys (Panels (a–d) and TMPRSS3 variant NP_076927.1:p.Met384Lys (Panels e–h). All hydrogen bonds are indicated by black dashed lines. a The wildtype HARS2 protein contains a tyrosine (blue) at position 364, which interacts with a neighboring cysteine amino acid (orange) b Augmentation of the boxed region in Panel A shows two hydrogen bonds between the tyrosine and cysteine. c The NP_036340.1:p.Tyr364Cys variant introduces a new cysteine (green) in place of tyrosine. d Enlargement of the boxed region from Panel C shows that the variant cysteine (green) interacts with the original neighboring cysteine (orange), disrupting the two hydrogen bonds to form a single hydrogen bond or a disulfide bond. e The wildtype TMPRSS3 protein shows a methionine (blue) at position 384, which interacts with three neighboring amino acids (orange). f Magnification of Panel E shows three hydrogen bonds between the methionine and neighboring amino acids. g The NP_076927.1:p.Met384Lys variant introduces a lysine (green) in place of methionine, which interacts with four neighboring amino acids, only one of which remains the same as the wildtype interacting neighbors. h Enlargement of the boxed region from Panel G shows four hydrogen bonds between the lysine (green) and neighboring amino acids. While one hydrogen bond remains the same between the wildtype and variant structures a black arrow indicates the residue with the unaltered hydrogen bond), the NP_076927.1:p.Met384Lys variant results in significant misfolding