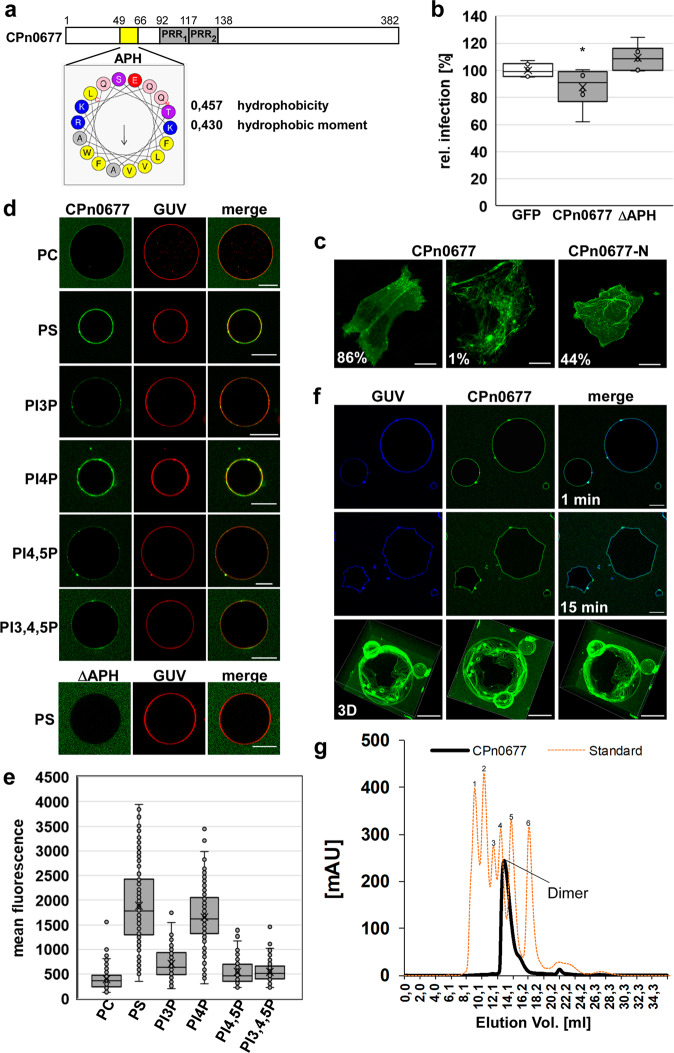
Fig. 1. The C-terminal segment of CPn0677 mediates membrane binding.
a Schematic representation of the domain structure of CPn0677. The position of the amphipathic helical region (APH, aa 49-66) is highlighted in yellow and was analyzed for hydrophobicity, hydrophobic moment and amino-acid composition using HeliQuest (heliquest.ipmc.cnrs.fr). Gray boxes represent proline-rich repeats (PRRs, aa 92–116; aa 117–138). b Quantification of inclusion formation in HEp-2 cells expressing either GFP, CPn0677 or CPn0677ΔAPH fused to GFP challenged with Cpn EBs (MOI1) 24 h post transfection. Inclusion numbers were quantified 48 hpi by staining the inclusion with an anti-LPS antibody and an anti-mouse antibody coupled to Alexa594. Inclusion numbers were normalized to those detected in GFP expressing control cells. ( ± SD, n = 3 biologically independent experiments) P = *≤ 0.05. c Confocal images of HEp-2 cells expressing CPn0677 or CPn0677-N fused to GFP. Bar: 10 µm. d Binding of FITC-labeled recombinant CPn0677 variants to GUVs containing the indicated lipids stained with Texas red and of GUVs incubated with CPn0677 and its N-terminal deletion derivative ΔAPH. Bar 10 µm. e Quantification of protein binding to GUVs containing the indicated lipids. Data are expressed as the mean fluorescence ( ± SD, n = 3 biologically independent experiments) of 50 GUVs shown in (d). f Confocal images of PS-GUVs labeled with Marina Blue™ and incubated with FITC-labeled CPn0677 (1 µM) for the indicated times (see also Supplementary Movie 1). Note the CPn0677-mediated membrane deformation at the 15-min timepoint. The bottom row shows 3D z-stack images taken from various angles at 15 min. Bar 10 µm. g SEC of 5.2 mg CPn0677 dissolved in PBS and separated on a superdex 200 increase 10/300 GL column depicted as solid black lane. The SEC profile of the molecular weights of the standard proteins is shown in orange dashed line (1Thyroglobulin 669 kDa, 2Apoferritin 480 kDa, 3β-Amylase 200 kDa, 4Alcohol Dehydrogenase 150 kDa, 5Albumin 66 kDa, 6Carbonic Anhydrase 29 kDa). mAU absorbance units. (n = 2 biologically independent experiments).