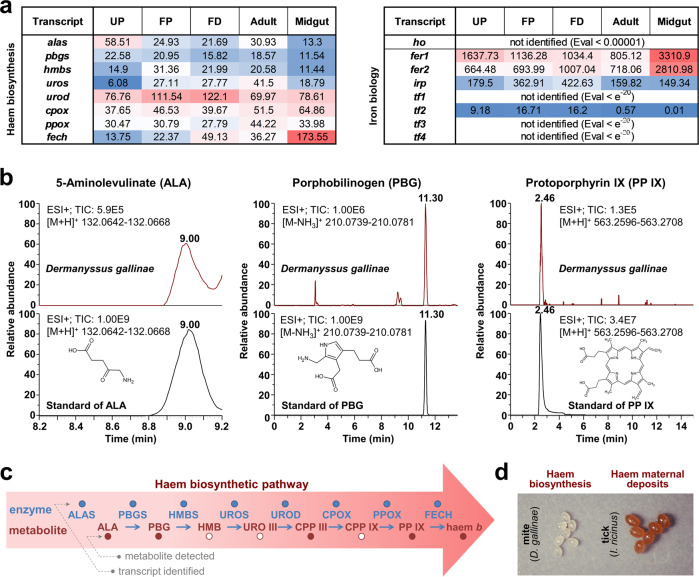
Fig. 4. Determination of full haem biosynthesis and identification of two distinct ferritin transcripts.
a Table of proteins participating in haem (left) or iron (right) homoeostasis, together with their expression values (FPKM) of their mRNA transcripts across libraries derived from individual developmental stages of Dermanyssus gallinae. Accession numbers are available in Supplementary Table S6. UP unfed protonymphs, FP fed protonymphs, FD fed deutonymphs. alas, 5-aminolevulinate synthase; pbgs, porphobilinogen synthase; hmbs, hydroxymethylbilane synthase; uros, uroporphyrinogen synthase; urod, uroporphyrinogen decarboxylase; cpox, coproporphyrinogen oxidase; ppox, protoporphyrinogen oxidase; fech, ferrochelatase; ho, haem oxygenase; fer, ferritin; tf, transferrin; irp, iron-responsive protein. b The LC/MS analysis of Haem b precursors in the naive non-fed stages of D. gallinae. Reconstructed chromatograms for the D. gallinae samples are shown on top, with reconstructed chromatograms for their analytical standards below. c A schematic of haem biosynthesis, which identifies (filled circles) transcripts and metabolites of the pathway. ALA δ-aminolevulinic acid, PBG porphobilinogen, HMB hydroxymethylbilane, URO uroporphyrinogen, CPP coproporphyrinogen, PP protoporphyrin. d A photographic image of eggs of D. gallinae mites and Ixodes ricinus ticks; note the colour difference, indicating the absence/presence of maternal haem deposits.