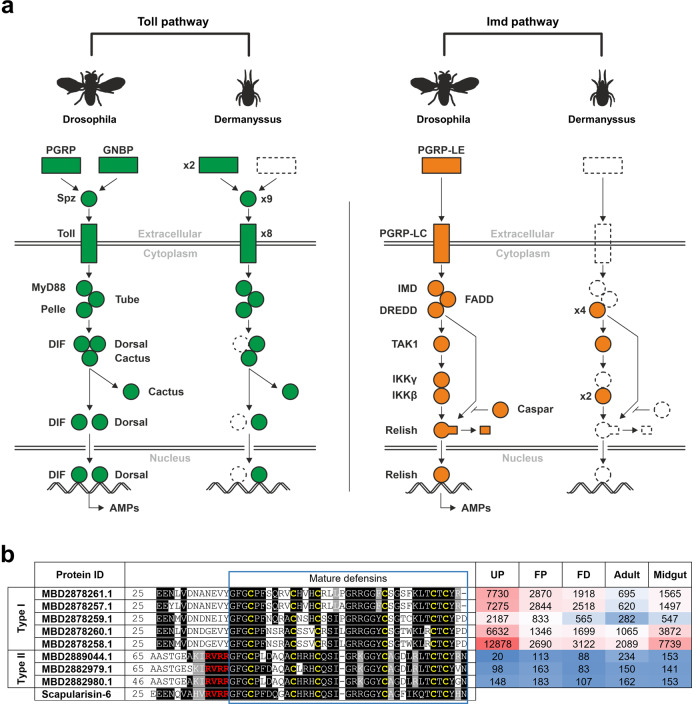
Fig. 7. Mining of immune pathways in the D. gallinae transcriptome.
a Protein sequences of Drosophila or Tribolium Toll and Imd pathways were used to search in our D. gallinae translated protein database (Local BLAST, E-value 0.1, Matrix BLOSUM62). Conservation of the domains was checked by CD-search (NCBI). Only homologous sequences with similar domain structures and an E-value < 10e−3 were considered putative homologues. Accession numbers are available as Supplementary Table S9. b Multiple sequence alignment of identified D. gallinae mature defensins, homology with tick Ixodes scapularis scapularisin-6 (GenBank EEC08935) and expression values (FPKM) of defensin transcripts across libraries derived from individual developmental stages of D. gallinae. The amino acid residues of the Furin cleavage site are shown in red letters. UP unfed protonymphs, FP fed protonymphs, FD fed deutonymphs.