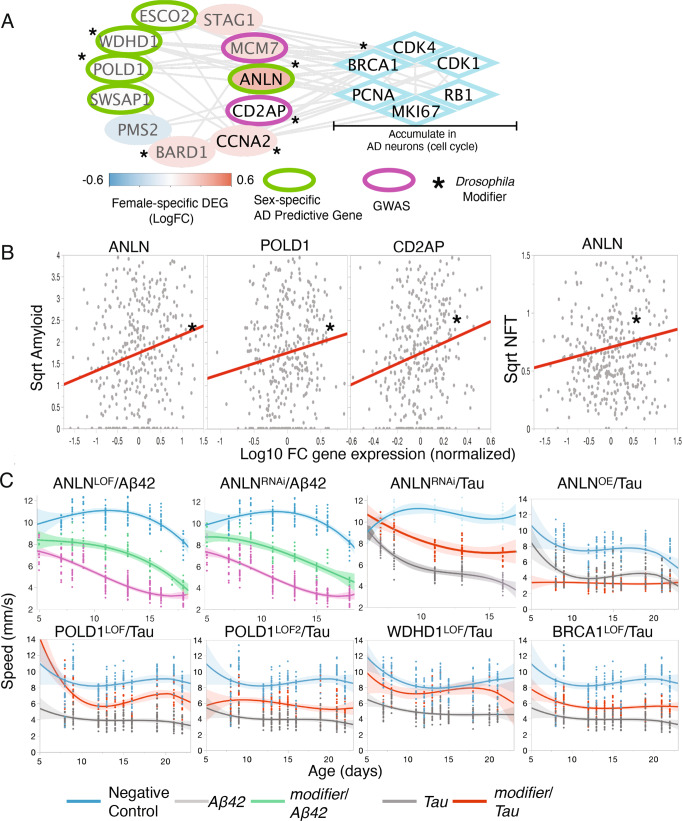
Fig. 4. Characterization of a cell-cycle/DNA repair-associated module enriched in female-specific EAML candidates.
A Integration of five female-specific EAML candidates involved in cell-cycle/DNA repair with genes predominantly dysregulated in female AD brains and cell cycle genes known to accumulate in AD neurons. B positive correlation of gene expression and neuropathologic features for some of the genes in the module shown in (A). C graphs representing longitudinal analyses of neuronal dysfunction assessed as speed of the animals as a function of age (days) for the indicated alleles. Blue corresponds to negative (healthy) controls expressing a non-targeting hp-RNA. Purple shows the performance of β42/ non-targeting and grey the performance of tau/ non-targeting hp-RNA diseased animals. Green or red show the performance of animals carrying the allele indicated on top and either expressing β42 (green) or tau (red) paneuronally. Knockdown of the Drosophila homologs of three of the EAML female-specific candidates (ANLN, POLD1, WDHD1) results in amelioration of the neurodegenerative phenotypes. Knockdown of the Drosophila homolog of BRCA1 also ameliorates neuronal dysfunction (specific alleles used are indicated in Supplementary Data 2). Each graph shows the fit curve of the third-degree polynomial regression (dark line) and the confidence intervals for the same regression (shaded). All experiments shown are statistically significantly different p < 0.05 (exact p values are shown in Supplementary Data 2) when analyzed using non-linear random mixed effects model ANOVA. Four replicates of ten animals each per genotype were used for these experiments.