Figure 2.
Trio vs. cohort analysis for expression outliers as well as aberrant junctions and allele-specific expression analysis in the cohort
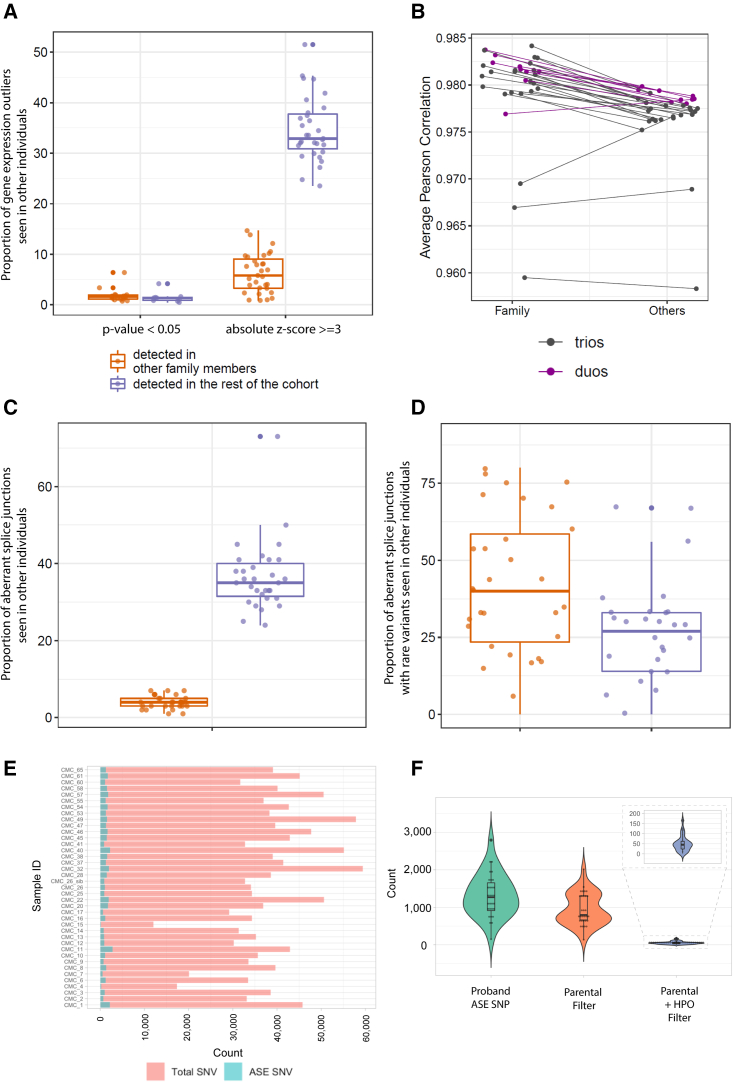
(A) Proportions of gene-expression outliers also detected in other samples showing that trio analysis is effective in filtering out statistically significant expression outliers, but not when using a more lenient cut-off (absolute Z score). Each dot represents a proband with family data available. y axis, proportion of gene-expression outlier defined by statistical significance (adjusted p value < 0.05) or Z scores (absolute Z score ≥ 3) that are also detected in family members (orange) or the rest of the cohort (purple).
(B) Gene expression correlation is consistently higher between probands and their family members than between probands and the rest of the cohort. y axis, average Pearson correlation of gene expression. Each dot represents a proband. Lines connect the same proband in the two columns. Purple, duos; black, trios.
(C) Proportions of total aberrant splicing events also detected in other samples. Each dot represents a proband with family data available. y axis, proportions of genes containing aberrant junctions detected in other family members (orange) or the rest of the cohort (purple).
(D) Proportions of aberrant splicing events with at least one rare variant nearby also detected in other samples. Each dot represents a proband with family data available. y axis, proportions of genes containing aberrant junctions detected in other family members (orange) or the rest of the cohort (purple).
(E) Bar plot of the number of reported SNVs and ASE SNVs for all the affected individuals. Red, total number of rare SNVs; blue, number of ASE events.
(F) Violin plot of the distribution of ASE SNVs after parental filter and HPO term filter.