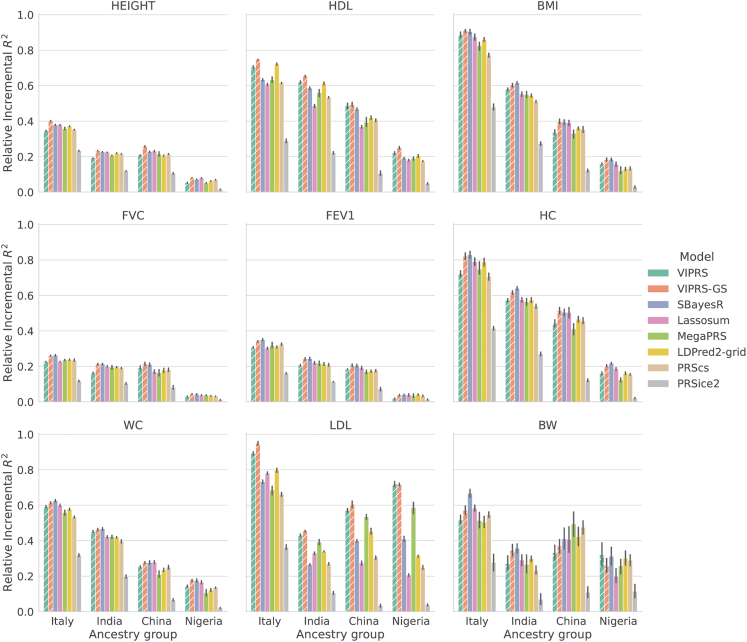
Figure 4.
Relative predictive performance of summary statistics-based PRS methods on real quantitative phenotypes in minority populations in the UK Biobank
The PRS models were trained on summary statistics from the White British cohort in the UK Biobank using a 5-fold cross-validation design. Then, the effect size estimates from the five training folds were used to perform predictions in individuals of Italian, Indian, Chinese, and Nigerian ancestry. Each panel shows the incremental prediction in a given ancestry group relative to the prediction of the best performing model on the White British cohort. The bars show the mean of the relative prediction metric across the five training folds and the black lines show the corresponding standard errors. The quantitative phenotypes analyzed are standing height (HEIGHT), high-density lipoprotein (HDL), body mass index (BMI), forced vital capacity (FVC), forced expiratory volume in 1 s (FEV1), hip circumference (HC), waist circumference (WC), low-density lipoprotein (LDL), and birth weight (BW). The PRS methods shown are our proposed VIPRS and VIPRS-GS (using grid search to tune model hyperparameters) as well as six other baseline models: SBayesR, Lassosum, MegaPRS, LDPred2 (grid), PRScs, and PRSice2 (C+T). Dashed lines highlight the models contributed in this work.