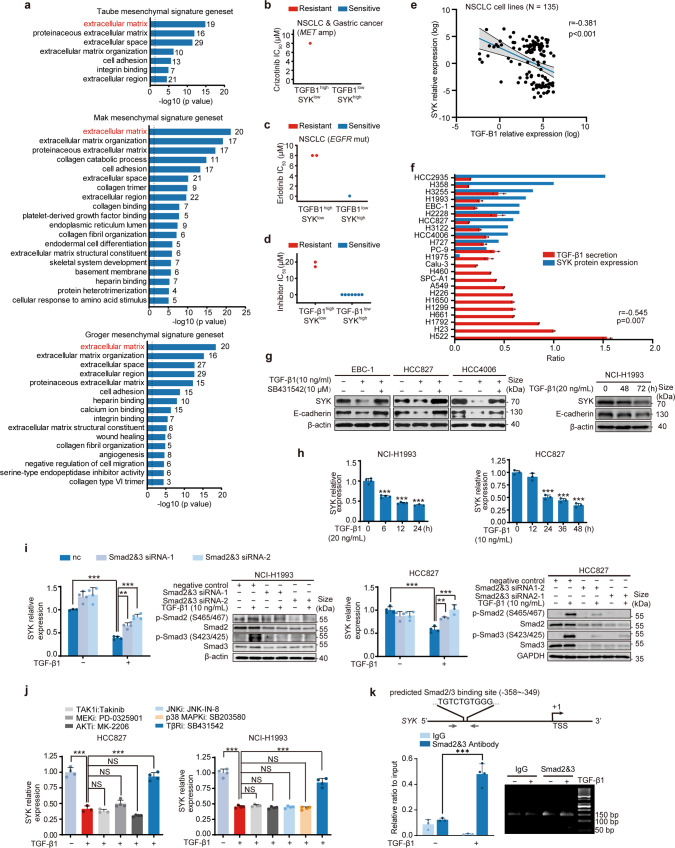
Fig. 3.
Transcriptional downregulation of SYK by TGF-β1 confers resistance to c-Metis and EGFRis. a GO annotation and enrichment analysis of mesenchymal signature gene sets (p < 0.05, FDR < 0.05). Genes in the extracellular matrix gene set are shown. b and c Samples were stratified based on crizotinib (b) or erlotinib (c) sensitivity according to the SYK/TGF-β1 mRNA expression pattern based on the data from CCLE. d A panel of available cancer cell lines (also seen in Fig. 1f) was stratified based on cell sensitivity to kinase inhibitors according to the SYK/TGF-β1 expression pattern. The secreted TGF-β1 level was normalized to the total cellular protein level. Samples were classified into high and low TGF-β1 subgroups using the median value as the cutoff in b-d. e Pearson correlation analysis between TGF-β1 and SYK mRNA expression in NSCLC cell lines based on the data from CCLE. f The association between TGF-β1 secretion and SYK protein levels in the profiled NSCLC cell lines. g Immunoblot analysis of the indicated cell lines treated with TGF-β1 alone or together with TGF-β1 inhibitor for 96 h or the indicated time. E-cadherin was investigated as a functional indicator of TGF-β1. h SYK mRNA level alterations in NCI-H1993 (left) or HCC827 (right) cells treated with TGF-β1. i, j Alteration of SYK transcription levels upon Smad2/3 knockdown (i) or treatment with the indicated inhibitors (j). k Smad2/3 enrichment in the SYK promoter measured by a ChIP assay. Upper: schematic diagram of SYK promoter showing location of putative Smad2/3 binding sites. The positions of the ChIP primers are delineated by gray arrowheads. TSS, transcription start site. Lower: qRT-PCR analysis (left) and agarose gel electrophoresis detection (right) by the ChIP assay. The data shown are representative results from two or three independent experiments in f–k. NS: no significance, ***p < 0.001, **p < 0.01, using Student’s t test in k, using one-way ANOVA in h, i, j