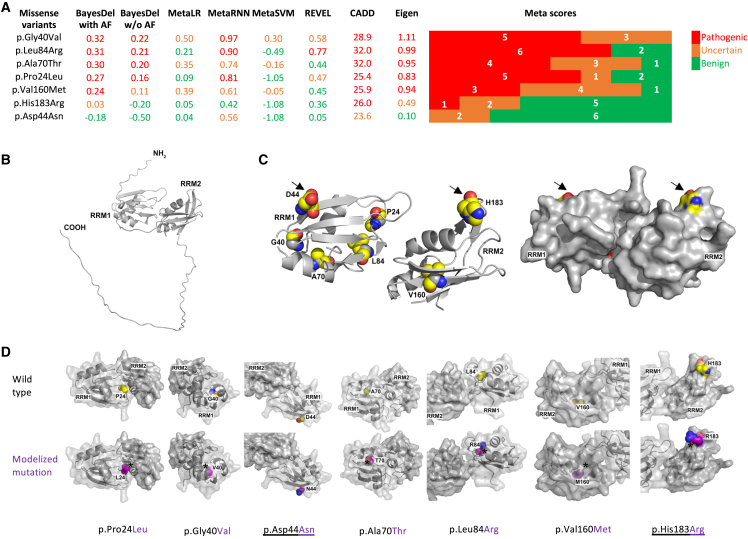
Figure 2.
Bio-informatic pathogenicity predictions and structural modeling of missense variants
(A) Pathogenicity prediction scores for the seven SRSF1 missense variants generated by eight meta-prediction tools (BayesDel with AF, BayesDel without AF, MetaLR, MetaRNN, MetaSVM, REVEL, CADD, Eigen) as implemented in the VarSome human genomic search engine.52 Colored bars on the right represent the number of meta-tools supporting pathogenic (red), uncertain (brown), and benign (green) predictions.
(B) Structural prediction of SRSF1 using AlphaFold and PyMol.
(C) Left: missense variants superimposed on the structural prediction. Right: Surface rendering of the SRSF1 protein structure. Arrows indicate p.Asp44Asn and p.His183Arg, two residues closer to the protein surface.
(D) Structural prediction of the seven missense variants. The underlined variants, p.Asp44Asn and p.His183Arg, are located at the protein surface. The other variants are more oriented toward the internal structure of the protein. Carbons are represented in yellow, nitrogens in blue, and oxygens in red for the wild-type amino acids; carbons are represented in purple in the modelized alterations. Asterisks indicate potential steric clashes in the mutated structures.