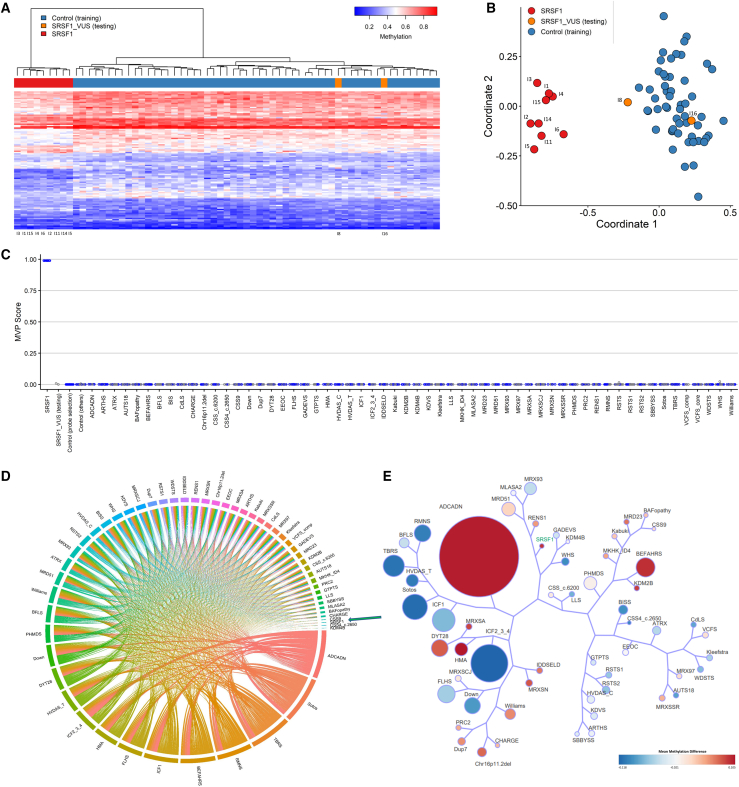
Figure 4.
Episignature assessment of SRSF1 VUSs p.Asp44Asn and p.His183Arg
(A) Heatmap indicates that the two VUS samples (orange) are clustering with controls (blue) and away from the SRSF1 samples with confirmed pathogenic variants (individuals I1–I6, I11, I14, and I15 used for episignature discovery) (red). Each row represents one of the 107 probes selected as the episignature, and each column represents an individual with either an SRSF1 variant (red or orange) or a control (blue).
(B) Multidimensional scaling plot (MDS) also shows clustering of the SRSF1 VUS samples with controls.
(C) Support vector machine classifier model (SVM) shows that the VUSs have a probability score (methylation variant pathogenicity score, MVP) of close to 0 compared with the SRSF1 samples carrying confirmed pathogenic variants with MVP scores of close to 1. The model is trained using the 107 selected SRSF1 episignature probes and 75% of controls and other neurodevelopmental disorder samples on EpiSign (blue circles). The 25% remaining are used as testing samples (gray circles).
(D) Circos plot representing the differentially methylated probes (DMPs) shared between each pair of cohorts. The thickness of the connecting lines indicates the number of probes shared between the paired cohorts. SRSF1 cohort is indicated by the green arrow.
(E) Tree-and-leaf visualization of Euclidean clustering of the SRSF1 cohort alongside the 56 other EpiSign disorders using the top n DMPs for each cohort, where n = 500 or the max number of DMPs available if <500. Cohort samples are aggregated using the median value of each probe within a group. Each leaf (node) represents a cohort, with node sizes illustrating relative scales of the number of selected DMPs for the corresponding cohort, and node colors indicative of the global mean methylation difference where blue is more hypomethylated and red hypermethylated. The SRSF1 cohort with confirmed pathogenic variants is highlighted in green. ADCADN, cerebellar ataxia deafness and narcolepsy syndrome; AUTS18, susceptibility to autism 18; BEFAHRS, Beck-Fahrner syndrome; BFLS, Borjeson-Forssman-Lehmann syndrome; BISS, blepharophimosis intellectual disability SMARCA2 syndrome; CdLS, Cornelia de Lange syndrome; CHARGE, CHARGE syndrome; Chr16p11.2del, chromosome 16p11.2 deletion syndrome; CSS, Coffin-Siris syndrome; CSS4, Coffin-Siris syndrome 4; CSS9, Coffin-Siris syndrome 9; Down, Down syndrome; Dup7, 7q11.23 duplication syndrome; DYT28, dystonia 28; EEOC, epileptic encephalopathy-childhood onset; FLHS, Floating-Harbor syndrome; GTPTS, genitopatellar syndrome; HMA, Hunter McAlpine craniosynostosis syndrome; HVDAS, Helsmoortel-van der Aa syndrome; ICF, immunodeficiency-centromeric instability-facial anomalies syndrome; IDDSELD, intellectual developmental disorder with seizures and language delay; Kabuki, Kabuki syndrome; KDVS, Koolen-De Vries syndrome; Kleefstra, Kleefstra syndrome; LLS, Luscan-Lumish syndrome; MKHK, Menke-Hennekam syndrome; MLASA2, myopathy lactic acidosis and sideroblastic anemia 2; MRD23, intellectual developmental disorder 23; MRD51, intellectual developmental disorder 51; MRX93, intellectual developmental disorder X-linked 93; MRX97, intellectual developmental disorder X-linked 97; MRXSA, intellectual developmental disorder X-linked syndromic Armfield type; MRXSCH, intellectual developmental disorder X-linked syndromic Christianson type; MRXSCJ, intellectual developmental disorder X-linked syndromic Claes-Jensen type; MRXSN, intellectual developmental disorder X-linked syndromic Nascimento type; MRXSSR, intellectual developmental disorder X-linked syndromic Snyder-Robinson type; PHMDS, Phelan-McDermid syndrome; PRC2, PRC2 complex (Weaver and Cohen-Gibson) syndrome; RENS1, Renpenning syndrome; RMNS, Rahman syndrome; RSTS, Rubinstein-Taybi syndrome; SBBYSS, Ohdo syndrome; Sotos, Sotos syndrome; TBRS, Tatton-Brown-Rahman syndrome; WDSTS, Wiedemann-Steiner syndrome; WHS, Wolf-Hirschhorn syndrome; Williams, Williams syndrome.