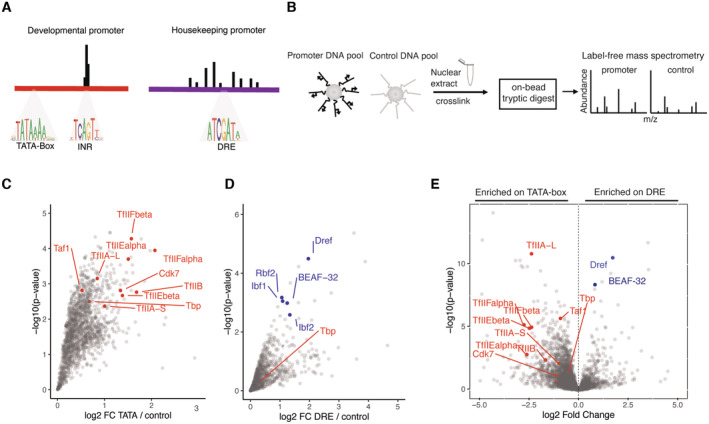
Figure 1. DNA affinity purifications uncover differentially bound proteins at functionally distinct promoters.
-
AExamples of stereotypical 121‐bp‐long core promoters used for DNA affinity purification of a developmental TATA‐box type, and a DRE motif‐containing housekeeping type. TATA‐box promoters exhibit focused transcription initiation from the INR motifs focused around 1–3 bp, while DRE‐containing promoters exhibit dispersed transcription initiation across 50–100 bp.
-
BScheme of DNA affinity purification coupled to label‐free mass spectrometry. Promoters are analyzed in pools and enrichment is measured against a pool of negative control regions from the Drosophila genome.
-
C, D(C) Enrichment of proteins detected by mass spectrometry on a pool of TATA‐box promoters over control DNA sequences, and in (D) over a pool of DRE promoters. Three biological replicates were performed for each promoter and control pool and significance measured with a Limma P‐value < 0.05.
-
EEnrichment of proteins bound to DRE promoters over TATA‐box promoters. Limma P‐value < 0.05. This comparison is generated by performing a ratio of abundance of proteins binding to DRE promoters directly over the abundance on TATA‐box promoters.
