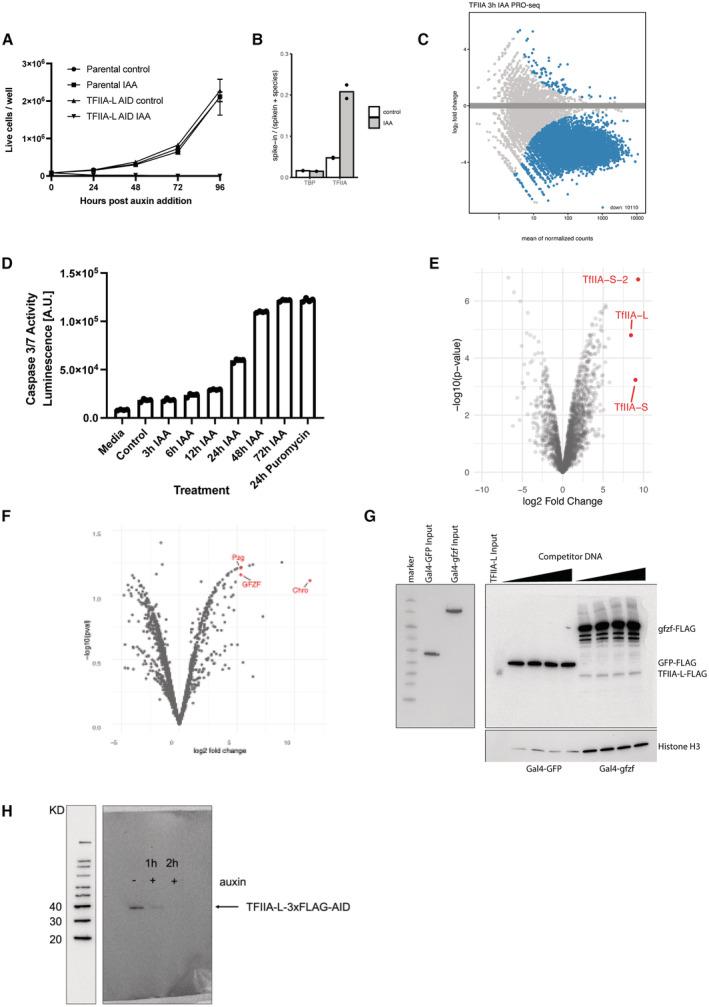
Growth curve of TFIIA‐L‐3xFLAG‐AID cell line and parental Tir1 expressing control over 4 days upon the addition of 500 μM auxin. TFIIA‐L‐AID treated cells die after 24 h. Error bars represent standard deviation across three biological replicates.
Fraction of reads mapping to the D. mel genome (reference species) and the human genome (spike‐in) in PRO‐seq experiments depleting TBP or TFIIA‐L. A ~ 4‐fold increase in proportion of reads mapping to the spike‐in genome is observed only upon depletion of TFIIA‐L due to global failure of Pol II transcription in the TFIIA‐L‐AID cell line.
MA‐plot of PRO‐seq data in the TFIIA‐L‐AID cell line after 3 h of auxin treatment, showing a global failure of Pol II transcription.
Caspase 3 and 7 activity was measured with the Promega Caspase 3/7 Glo kit of TFIIA‐L‐AID cells after addition of auxin at various time points. A positive cell death control was included as a 24 h treatment of 10 μg/ml puromycin.
Volcano plot of TFIIA‐L immunoprecipitation mass spectrometry. TFIIA‐L was immunoprecipitated from the endogenously tagged TFIIA‐L‐3xFLAG‐AID cell line using anti‐FLAG beads. Enrichment was measured over control immunoprecipitation made from the Tir1 expressing parental cell line which does not contain any FLAG epitope. Three replicates were performed for each condition.
Volcano plot of Chromator immunoprecipitation mass spectrometry. Chromator was immunoprecipitated from the Chromator‐3xFLAG‐AID cell line using an anti‐FLAG antibody. Similar Tir1 expressing parental cell line control was used to measure enrichment. Putzig (Pzg) and GFZF are also highlighted.
DNA affinity purification assay was performed with a 121‐bp‐long housekeeping DRE promoter with 4xUAS sites upstream. Initially, a nuclear extract containing a Gal4‐DNA‐binding domain fusion of GFP or GFZF was incubated with the bead‐immobilized promoter DNA (left panel). After the incubation, the extract was removed, and the beads were used for a DNA affinity purification assay with a nuclear extract containing TFIIA‐L‐AID‐3xFLAG as described in the materials and methods. Sheared salmon sperm DNA was used as competitor DNA at 600 ng to 1.6 μg per reaction. Elution fractions were run on an SDS–PAGE gel and blotted with a FLAG antibody (right panel).
Western blot against FLAG antibody visualizing whole cell lysate from a TFIIA‐L c‐terminally tagged 3x‐FLAG‐AID line treated with auxin for 2 h. Full degradation of the TFIIA‐L beta subunit is visible upon 2 h of auxin treatment.