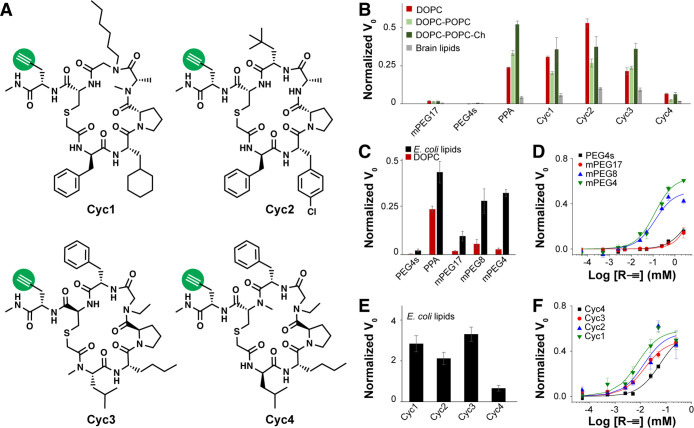
Figure 2.
Liposomal permeation assay using different lipid mixtures. (A) Various cell-permeable macrocycles were alkyne-labeled (Cyc1–4). (B) Permeation rates of mPEG17, PEG4s, PPA, and Cyc1–4 were measured using liposomes composed of DOPC, DOPC/POPC, DOPC/POPC/Ch (20 mol % Ch), and brain polar lipids. Initial CuAAC reaction velocity (V0, fold/min) in the liposomal assay was normalized to the corresponding reaction velocity without liposomes. (C) mPEG17, PEG4s, PPA, mPEG8, and mPEG4 were tested in liposomal permeation assays of E. coli extract polar lipids and DOPC, and (D) concentration-dependent permeation was measured. (E) Cyc1–4 were tested in liposomal permeation assays of E. coli extract polar lipids, and further measured at varied concentrations in the assay (F). Error bars indicate the standard error of the mean (N = 3).