Abstract
Objective
The incidence of stroke has been rising annually and investigations into traditional risk factors have led to increased attention on genetic factors. In this study, we focused on the pri-let-7f gene, and investigated the association between pri-let-7f gene polymorphisms and ischemic stroke (IS).
Methods
This case–control study included 1803 patients and 1456 healthy controls of Han ethnicity living in Liaoning Province. We carried out genotyping analysis of two loci, pri-let-7f-1 rs10739971 and pri-let-7f-2 rs17276588, and performed statistical analysis controlling for confounding factors by logistic regression.
Results
The A alleles and AA genotypes of both loci were significantly associated with an increased risk of IS. Variant genotypes of rs17276588 may also increase the risk of IS in females with alcohol intake. Gene–gene interaction analysis showed combined effects of mutations in both these single nucleotide polymorphisms (SNPs).
Conclusions
This study demonstrated an association between pri-let-7f SNPs and IS, providing potential latent biomarkers for the risk of IS. However, more detailed studies are needed to clarify these results.
Keywords: Single nucleotide polymorphism, pri-microRNA, pri-let-7f, ischemic stroke, gene–environment interaction, gene–gene interaction
Introduction
Stroke is a leading cause of death globally and has a significant impact on human health, with high morbidity and an annual mortality of 5.5 million. 1 Almost 50% of stroke patients have to live with a chronic handicap for the rest of their life, and cerebrovascular disease is the second-leading cause of disability-adjusted life years. 2 China has the highest lifetime risk of stroke, estimated as 39.9%. 3 The prevalence of stroke in China has increased annually from 2013 to 2019, especially in northeast China, 4 and the high disability rate and high lifetime risk, together with the high prevalence, place a financial burden on society and physical and psychological burdens on caregivers. Stroke can be classified as ischemic stroke (IS) or hemorrhagic stroke, with IS accounting for around 80% of all stroke cases. 5 There is thus an urgent need to identify the mechanism underlying IS, to improve stroke-prevention strategies, treatment methods, and prognosis. Traditional risk factors for IS, such as age, sex, and hypertension, fail to explain stroke risk adequately 6 ; however, genetic variables have been shown to participate in the pathophysiology of neurological diseases,7,8 and increasing evidence suggests that genes may play a potential role in the pathophysiology of IS.
MicroRNAs (miRNAs) are endogenous non-coding RNAs that regulate mRNAs to affect their translation.9,10 MiRNAs are initially transcribed as hairpin-containing primary transcripts (pri-miRNAs) by RNA polymerase II and then processed into precursor miRNAs (pre-miRNAs). 11 Previous studies have demonstrated the functional influence of pri-miRNAs on miRNAs; e.g., methyltransferase 1-mediated methylation regulated the structure of let-7 by changing the structure within the pri-miRNA. 12 Let-7, the first known human miRNA, is related to stem cell division and differentiation. Its family consists of 10 members, designated a–j respectively, derived from 13 precursor sequences. Let-7 miRNAs have highly conserved sequences and functions. 13 Single nucleotide polymorphisms (SNPs) have been suggested to act as risk factors in some diseases, 14 and the association between SNPs and IS has been explored using genome-wide association studies and candidate-gene studies.15–19 However, these explorations are complicated by the need for accurate selection and verification from among large numbers of potential SNPs, as well as the complex pathogenesis of IS and the difficulty in avoiding racial factors. Nevertheless, it is important to continue such studies, with suitable precautions, especially in light of developing methodologies.
Researchers found that pri-let-7 SNPs may affect the expression of mature let-7, and numerous studies have investigated the relationship between pri-let-7 SNPs and cancers.20,21 Let-7f exerts functions in cerebral ischemia by targeting NDRG3, indicating its role in IS. 22 Two SNPs in the primary precursor area of the let-7f family (pri-let-7f-1 rs10739971, pri-let-7f-2 rs17276588) have been associated with cancers,20,23 confirming the relevance of pri-let-7f and let-7f; however, information on the association between these pri-let-7f SNPs and IS is still lacking.
In this study, we conducted a case–control study of 3259 subjects from northeast China, including 1803 patients with IS and 1456 healthy controls. We analyzed the two chosen SNPs, pri-let-7f-1 rs10739971 and pri-let-7f-2 rs17276588, using dominant and recessive models. The locus rs17276588 is located on the X chromosome, resulting in a sex difference. We determined the additive and multiplicative effects and explored the gene–gene and gene–environment interactions using logistic regression. We aimed to clarify the latent link between these two SNPs and IS, to further our understanding of primary IS.
Materials and methods
Subjects and design
This case–control study included patients with IS and age- and sex-matched healthy controls. Patient data were collected from the First Affiliated Hospital of China Medical University from December 2013 to December 2018. Eligible patients were first diagnosed with acute IS according to the following criteria: (1) abrupt onset of focused neurological deficits; (2) deficits that lasted longer than 24 hours; and (3) available brain imaging of the infarction. Patients with transient ischemic attack, cardioembolism, brain trauma, cerebrovascular malformations, coagulation dysfunction, autoimmune disorders, malignancies, chronic infectious diseases, or diseases of other systems were excluded from the study. Controls were recruited from individuals who underwent physical examination at the First Affiliated Hospital of China Medical University, with no evidence of stroke or other neurological illnesses.17–19,24 Body mass index (BMI) was classified in accordance with the Asian obesity classification, and a BMI >22.9 was considered overweight. Hyperlipidemia was defined according to current criteria (triglycerides >1.7 mmol/L or/and total cholesterol >5.72 mmol/L).
This study was authorized by the Institutional Ethics Committee of China Medical University First Hospital on 20 February 2012 (No. 2012-38-1), and performed according to the World Medical Association’s Code of Ethics (Declaration of Helsinki). All the participants provided written informed consent and their details were de-identified for privacy. The protocol was recorded in the Chinese Clinical Trial Registry (registration number: ChiCTR-COC-17013559) on 27 December 2017. The reporting of this study conforms to the Genetic Risk Prediction Studies (GRIPS) guidelines. 25
SNP selection
SNPs were searched using the UCSC genome browser (http://genome.ucsc.edu/) and dbSNP database (https://www.ncbi.nlm.nih.gov/snp/?term=dbSNP). We identified two SNPs in the primary precursor region of let-7f (pri-let-7f-1 rs10739971, pri-let-7f-2 rs17276588) with a minor allele frequency (MAF). SNPs with a MAF >5% in the Chinese population were all tagSNPs, but their potential predicting roles were unclear. We selected these two SNPs on the basis of the following criteria: (1) located in the region −1 kb upstream of pri-let-7f; and (2) MAF >0.1 in Han Chinese. The two SNPs were rs10739971, located in the −949 base pairs upstream of pri-let-7f-1, and rs17276588 located in the −183 base pairs upstream of pri-let-7f-2 (Table 1).
Table 1.
Characteristics of the let-7f single nucleotide polymorphisms selected for study.
| SNP ID | Gene | Chromosome | HWEp value | Genomic location | Alleles (major/minor) | MAF |
|---|---|---|---|---|---|---|
| rs10739971 | pri-let-7f-1 | 9 | 0.299 | 96937680 | G/A | 0.389 |
| rs17276588 | pri-let-7f-2 | X | 0.952 | 53601143 | G/A | 0.274 |
SNP, single nucleotide polymorphism; HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency.
DNA extraction and genotyping
Genomic DNA was extracted from ethylenediaminetetraacetic acid-anticoagulated peripheral blood and stored at −80°C. The SNaPshot reaction was performed, as described previously.17,18 DNA was extracted using a DNA Purification Kit (Promega, Madison, WI, USA) and genotype analysis was carried out using a SNaPshot Multiplex Kit (Applied Biosystems Co., Ltd., Foster City, CA, USA). The primer sequences used for polymerase chain reaction are shown in Table 2. The results were analyzed using an ABI 3130XL DNA sequence detector and GeneMapper 4.0 (Applied Biosystems Co., Ltd.).
Table 2.
Primer sequences.
| SNP | Primer sequence | Product size (bp) |
|---|---|---|
| rs10739971 | Forward: 5′-TGGACTCTGCCTTCAATCCACAT-3′ | 221 |
| Reverse: 5′-CATCATGCATAATCCAAATGCACTAAC-3′ | ||
| rs17276588 | Forward: 5′-TCAGCCTATGTGGGCCAGCTAC-3′ | 227 |
| Reverse: 5′-GATGGTCTGGGTGGAGGATGGT-3′ |
SNP, single nucleotide polymorphism; bp, base pairs.
Statistical analyses
All statistical analyses were performed using SPSS v23.0 (IBM Corp., Armonk, NY, USA), unless otherwise noted. All tests were two-tailed and significance was defined as p < 0.05. We compared the distributions of demographic variables using Pearson’s χ2 test and examined differences between risk factors and genotypes for alleles between patients and controls. A goodness-of-fit χ2 test was used to test the Hardy–Weinberg equilibrium for each genotype. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated by unconditional logistic regression to estimate the association between IS and a particular genotype. The QUANTO power calculator was used to calculate the power in a cohort. Assuming a genotypic relative risk for the dominant model of 2, a MAF of 0.3, a 1.12% population prevalence of IS, and a Type I error probability of 0.05 in a sample size of 1803 patient samples and 1456 healthy controls, we would be able to reject the null hypothesis that OR = 1 with a power of 99.99% with a Type II error probability of 0.0001.
The attributable proportion due to interaction (AP) and relative excess risk due to interaction (RERI) were used to test additive gene–gene and gene–environment interactions. If RERI and AP = 0, there was no biologic interaction. 26 Gene interactions between the two SNPs were evaluated under additive and multiplicative models, using a calculator in Excel (available at http://www.epinet.se). 27 Confounding factors including sex, age, BMI, hypertension, diabetes mellitus, history of alcohol use, history of smoking, family history, and hyperlipidemia were controlled in the logistic regression analysis.
Results
Characteristics of study subjects
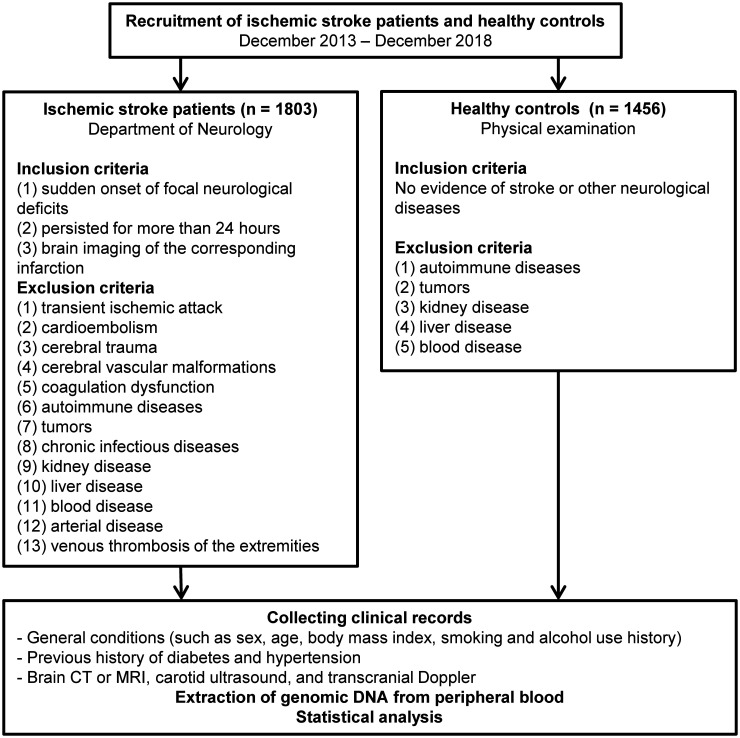
The study enrolled 1803 patients with IS and 1456 age - and sex-matched healthy controls, all of Han ethnicity, living in Liaoning Province in northern China. All patients were aged 40 to 80 years old. The study flow chart is shown in Figure 1 and the essential characteristics of the selected cases and controls, as well as the risk factors of IS, are summarized in Table 3. There was no significant difference between the cases and controls in terms of age or sex, but BMI, diabetes mellitus, hypertension, smoking history, family history, history of alcohol use, and hyperlipidemia all differed significantly between the two groups (p < 0.001), in accordance with conventional risk factors. Patients tended to have higher total cholesterol, triglyceride, and low-density lipoprotein cholesterol levels, and lower high-density lipoprotein levels than controls. The patients were age- and sex-matched, implying that the sample was representative.
Figure 1.
Flowchart of this study.
CT, computed tomography; MRI, magnetic resonance imaging.
Table 3.
Characteristics and risk factors for stroke
| Variable | Cases (%) | Controls (%) | p value |
|---|---|---|---|
| Age (≤60/>60 years) | 1083 (60.1)/720 (39.9) | 921 (63.3)/535 (36.7) | 0.065 |
| Sex (male/female) | 1032 (57.2)/771 (42.8) | 826 (56.7)/630 (43.3) | 0.776 |
| BMI (≤22.9/>22.9) | 939 (52.1)/864 (47.9) | 844 (58.0)/612 (42.0) | 0.01 |
| Diabetes mellitus | 501 (27.8)/1302 (72.2) | 268 (18.4)/1188 (81.6) | < 0.001 |
| Hypertension | 1136 (63.0)/667 (37.0) | 623 (42.8)/833 (57.2) | < 0.001 |
| Preexisting medication (aspirin) | 194 (10.8)/1609 (89.2) | – | – |
| Preexisting medication (statins) | 99 (5.5)/1704 (94.5) | – | – |
| Thrombolytic therapy | 458 (25.4)/1345 (74.6) | – | – |
| Family history | 134 (7.4)/1669 (92.6) | 46 (3.2)/1410 (96.8) | < 0.001 |
| History of smoking | 750 (41.6)/1053 (58.4) | 425 (29.2)/1031 (70.8) | < 0.001 |
| History of alcohol use | 362 (20.1)/1441 (79.9) | 208 (14.3)/1248 (85.7) | < 0.001 |
| Hyperlipidemia | 820 (45.5)/983 (54.5) | 586 (40.2)/870 (59.8) | < 0.001 |
BMI, body mass index.
Genetic polymorphisms and effects on stroke risk
The genotype distributions of the two polymorphisms in IS cases and controls complied with the Hardy–Weinberg equilibrium. The frequency of rs10739971 genotype AA was significantly higher in patients (21.3%) compared with controls (17.0%) (OR = 1.330, CI = 1.081–1.637, p = 0.007) (Table 4). We further analyzed the frequencies in dominant (OR = 1.173, CI = 1.007–1.367, p = 0.041) and recessive models (OR = 1.246, CI = 1.037–1.497, p = 0.019). These results indicated that genotype AA might be associated with an increased risk of IS. Allele analysis also showed a significant difference in the frequency of the A allele (OR = 1.189, CI = 1.077–1.312, p < 0.001), suggesting that the A allele of rs10739971 was a possible risk factor for IS.
Table 4.
Allele and genotype frequencies of genetic polymorphisms among cases and controls and their main effects on stroke risk.
| SNP | Cases | % | Controls | % | OR (95% CI)*a | P valueb |
|---|---|---|---|---|---|---|
| rs10739971 genotype | ||||||
| GG (ref) | 549 | 30.4 | 505 | 34.7 | 1.00 (ref) | |
| GA | 870 | 48.3 | 703 | 48.3 | 1.116 (0.949–1.313) | 0.185 |
| AA | 384 | 21.3 | 248 | 17.0 | 1.330 (1.081–1.637) | 0.007 |
| Dominant effect | ||||||
| GG (ref) | 549 | 30.4 | 505 | 34.7 | 1.00 (ref) | |
| GA + AA | 1254 | 69.6 | 951 | 65.3 | 1.173 (1.007–1.367) | 0.041 |
| Recessive effect | ||||||
| GA + GG (ref) | 1419 | 78.7 | 1208 | 83.0 | 1.00 (ref) | |
| AA | 384 | 21.3 | 248 | 17.0 | 1.246 (1.037–1.497) | 0.019 |
| rs10739971 allele | ||||||
| G (ref) | 1968 | 54.6 | 1713 | 58.8 | 1.00 (ref) | |
| A | 1638 | 45.4 | 1199 | 41.2 | 1.189 (1.077–1.312) | < 0.001 |
| rs17276588 genotype | ||||||
| GG (ref) | 879 | 48.8 | 1180 | 81.0 | 1.00 (ref) | |
| GA | 366 | 20.3 | 129 | 8.9 | 4.181 (3.260–5.362) | < 0.001 |
| AA | 558 | 30.9 | 147 | 10.1 | 4.257 (3.437–5.273) | < 0.001 |
| Dominant effect | ||||||
| GG(ref) | 879 | 48.8 | 1180 | 81.0 | 1.00 (ref) | |
| GA + AA | 924 | 51.2 | 276 | 19.0 | 4.225 (3.579–4.987) | < 0.001 |
| Recessive effect | ||||||
| GA + GG (ref) | 1245 | 69.1 | 1309 | 89.9 | 1.00 (ref) | |
| AA | 558 | 30.9 | 147 | 10.1 | 4.008 (4.233–4.969) | < 0.001 |
| rs17276588 allele | ||||||
| G (ref) | 2464 | 68.3 | 2436 | 83.7 | 1.00 (ref) | |
| A | 1142 | 31.7 | 476 | 16.3 | 4.106 (3.632–4.641) | < 0.001 |
*ORs and 95% CIs calculated by logistic regression.
Adjusted OR(95%CI) and p value adjusted for age, sex, body mass index, diabetes mellitus, hypertension, history of smoking, history of alcohol use, family history and hyperlipidemia.
SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Regarding the rs17276588 pri-let-7f-2 polymorphism, the genotype frequencies of the GG, GA, and AA genotypes in the patients were 48.8%, 20.3%, 30.9%, respectively, compared with 81.0%, 8.9%, and 10.1% among the controls, with significant differences between patients and controls for AA (OR = 4.257, CI = 3.437–5.273, p < 0.001) and GA (OR = 4.181, CI = 3.260–5.362, p < 0.001) (Table 4). The results of the dominant and recessive models also indicated that genotypes AA and GA might be risk factors for IS. The frequency of the A allele differed significantly between the groups (OR = 4.106, CI = 3.632–4.641, p < 0.001), suggesting that the A allele of rs17276588 may be a risk factor for IS.
Given that rs17276588 is located on the X chromosome, we analyzed its allele and genotype frequencies in both sexes (Table 5). No GA genotype was found in men because men only have a single X chromosome. The A allele frequency differed significantly between patients and controls in men (OR = 5.032, CI = 4.296–5.894, p < 0.001), in accordance with the genotype analysis, showing an increased risk for AA compared with GG (OR = 4.403, CI = 3.475–5.578, p < 0.001). Similar to the overall population, the AA genotype differed between patients and controls in women (OR = 3.929, CI = 2.207–6.992, p < 0.001), but the OR in men was obviously higher than that for the whole population, making the AA genotype a greater risk factor for IS in the male population. The outputs of the dominant (OR = 3.920, CI = 3.094–4.966, p < 0.001) and recessive (OR = 2.448, CI = 1.385–4.329, p = 0.002) models also suggested that AA genotype might be a risk factor and the A allele might be a risk allele (OR = 3.060, CI = 2.514–3.725, p < 0.001).
Table 5.
Allele and genotype frequencies of genetic polymorphisms among cases and controls and their main effects on stroke risks in both sexes.
| Sex | SNP | Cases | % | Controls | % | OR (95% CI)*a | p valueb | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Male | rs17276588 genotype | |||||||||
| GG (ref) | 532 | 51.6 | 696 | 84.3 | 1.00 (ref) | |||||
| AA | 500 | 48.4 | 130 | 15.7 | 4.403 (3.475–5.578) | <0.001 | ||||
| rs17276588 allele | ||||||||||
| G (ref) | 1064 | 51.6 | 1392 | 84.3 | 1.00 (ref) | |||||
| A | 1000 | 48.4 | 260 | 15.7 | 5.032 (4.296–5.894) | <0.001 | ||||
| Female | rs17276588 genotype | |||||||||
| GG(ref) | 347 | 45.0 | 484 | 76.8 | 1.00 (ref) | |||||
| GA | 366 | 47.5 | 129 | 20.5 | 3.919 (3.061–5.016) | <0.001 | ||||
| AA | 58 | 7.5 | 17 | 2.7 | 3.929 (2.207–6.992) | <0.001 | ||||
| Dominant effect | ||||||||||
| GG (ref) | 347 | 45.0 | 484 | 76.8 | 1.00 (ref) | |||||
| GA+AA | 424 | 55.0 | 146 | 23.2 | 3.920 (3.094–4.966) | <0.001 | ||||
| Recessive effect | ||||||||||
| GA+GG (ref) | 713 | 92.5 | 613 | 97.3 | 1.00 (ref) | |||||
| AA | 58 | 7.5 | 17 | 2.7 | 2.448 (1.385–4.329) | 0.002 | ||||
| rs17276588 allele | ||||||||||
| G (ref) | 1060 | 68.7 | 1097 | 87.1 | 1.00 (ref) | |||||
| A | 482 | 31.3 | 163 | 12.9 | 3.060 (2.514–3.725) | <0.001 | ||||
ORs and 95% CIs calculated by logistic regression.
Adjusted OR(95%CI) and p value, adjusted for age, body mass index, diabetes mellitus, hypertension, history of smoking, history of alcohol use, family history and hyperlipidemia.
SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Gene–environment interactions
We analyzed the associations between genetic polymorphisms and risk-factor exposure. There was no multiplicative interaction between the rs17276588 polymorphism in either sex and a history of alcohol use (Table 6), but there was an additive effect of these two factors in the overall population, with a positive RERI and an almost qualified AP (RERI = 2.164, CI = 0.283–4.045; AP = 0.301, CI = −0.026 to 0.627). Individuals meeting both conditions were more susceptible to IS, indicating an overall synergistic effect between rs17276588 polymorphism and a history of alcohol use. The results stratified by alcohol exposure (Table 7) suggested that the significant association depended more on the polymorphism (wild-type or not) than on alcohol status in the female population (OR = 3.766, CI = 2.906–4.879, p < 0.001), but alcohol use increased the risk in women (OR = 5.345, CI = 3.144–9.085, p < 0.001). The GA and AA genotypes differed significantly between patients and controls in both men and women, irrespective of alcohol use, supporting the possibility that the A allele was a risk factor for IS.
Table 6.
Multiplicative model for interaction between genetic polymorphisms and history of alcohol use on risk of ischemic stroke.
| Genotype | Risk factor exposure | Male |
Female |
||
|---|---|---|---|---|---|
| OR (95% CI)* | p value | OR (95% CI)* | p value | ||
| rs10739971 | History of alcohol use | 0.744 (0.434–1.275) | 0.282 | 0.751 (0.420–1.340) | 0.332 |
| rs17276588 | 1.136 (0.594–2.171) | 0.700 | 0.770 (0.409–1.448) | 0.417 | |
ORs and 95% CIs calculated by logistic regression.
OR, odds ratio; CI, confidence interval.
Table 7.
Association between rs17276588 polymorphisms and risk of ischemic stroke stratified by alcohol exposure in both sexes.
| rs17276588 genotype | Alcohol status | Male |
Female |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases (%) | Controls (%) | OR (95%CI)*a | p valueb | Cases (%) | Controls (%) | OR (95%CI)*a | p valueb | ||
| GG | No | 422 (40.9) | 611 (74.0) | 1.00 (ref) | 274 (35.5) | 397 (63.0) | 1.00 (ref) | ||
| GA + AA | No | 406 (39.3) | 114 (13.8) | 6.156 (3.457–10.963) | < 0.001 | 339 (44.0) | 126 (20.0) | 3.766 (2.906–4.879) | < 0.001 |
| GG | Yes | 110 (10.7) | 85 (10.3) | 1.505 (1.064–2.129) | 0.021 | 73 (9.5) | 87 (13.8) | 1.119 (0.773–1.618) | 0.551 |
| GA + AA | Yes | 94 (9.1) | 16 (1.9) | 4.456 (3.454–5.749) | < 0.001 | 85 (11.0) | 20 (3.2) | 5.345 (3.144–9.085) | < 0.001 |
ORs and 95% CIs calculated by logistic regression.
Adjusted OR(95%CI) and p value adjusted for age, body mass index, diabetes mellitus, hypertension, history of smoking, family history and hyperlipidemia.
OR, odds ratio; CI, confidence interval.
Gene–gene interactions
We determined the additive and multiplicative effects of the SNPs in both sexes using a dominant model (Table 8), and showed that the two SNPs had a synergistic interaction. There were significant interactions in the multiplicative model in both men (OR = 1.624, CI = 1.022–2.580, p = 0.040) and women (OR = 2.155, CI = 1.269–3.663, p < 0.001), but the interaction was more significant and the OR was higher in women, suggesting a greater effect in women. There were also significant interactions between the polymorphisms in both men (OR = 2.705, CI = 1.979–3.696, p < 0.001) and women (OR = 4.610, CI = 3.322–6.397, p < 0.001) in the additive model. The results of the dominant model separated by sex may account for the sex differences in the multiplicative model.
Table 8.
Multiplicative and additive models of association between two single nucleotide polymorphisms and risk of ischemic stroke.
| SNP | Male |
Female |
||
|---|---|---|---|---|
| OR (95%CI)* | p value | OR (95%CI)* | p value | |
| rs10739971 (GA or AA vs GG) | 1.561 (1.048–2.325) | 0.028 | 0.810 (0.526–1.247) | 0.338 |
| rs17276588 (GA or AA vs GG) | 5.978 (4.500–7.941) | < 0.001 | 3.238 (2.463–4.256) | < 0.001 |
| Multiplicative model | 1.624 (1.022–2.580) | 0.040 | 2.155 (1.269–3.663) | < 0.001 |
| Additive model | 5.746 (4.207–7.849) | < 0.001 | 5.652 (4.028–7.932) | < 0.001 |
*ORs and 95% CIs calculated by logistic regression.
SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
The combined effects of the two polymorphisms in men and women are shown in Table 9. In men, mutated rs17276588 genotypes combined with either wild-type GG (OR = 2.962, CI = 2.009–4.366, p = 0.028) or mutated GA+AA (OR = 4.828, CI = 3.481–6.695, p < 0.001) in rs10739971 were all significant and the rs10739971 A allele had a higher OR, while any rs10739971 genotype combined with wild-type rs17276588 was not significant. In women, any single mutation in either SNP affected the risk of IS. From Table 10, stratified by rs10739971, the OR was >1 in both sexes, with a higher IS risk in females than in males; when stratified by rs17276588, mutated types in males (OR = 1.584, CI = 1.017–2.469, p = 0.042) and wild-type in females (OR = 1.705, CI = 1.232–2.359, p < 0.001) were significant in a dominant model of rs10739971, and the relatively small p value indicated a meaningful trend for GA+AA in females.
Table 9.
Combined effects of rs10739971 and rs17276588 polymorphisms on ischemic stroke risk.
| rs10739971 | rs17276588 | Male |
Female |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases (%) | Controls (%) | OR (95%CI)*a | p valueb | Cases (%) | Controls (%) | OR (95%CI)*a | p valueb | ||
| GG | GG | 189 (18.3) | 241 (29.2) | 1.00 (ref) | 85 (11.0) | 175 (27.8) | 1.00 (ref) | ||
| GA + AA | GG | 343 (33.2) | 455 (55.1) | 0.889 (0.692–1.142) | 0.357 | 262 (34.0) | 309 (49.0) | 1.680 (1.228–2.297) | 0.001 |
| GG | GA + AA | 153 (14.8) | 53 (6.4) | 2.962 (2.009–4.366) | < 0.001 | 122 (15.8) | 36 (5.7) | 6.792 (4.286–10.763) | < 0.001 |
| GA + AA | GA + AA | 347 (33.6) | 77 (9.3) | 4.828 (3.481–6.695) | < 0.001 | 302 (39.2) | 110 (17.5) | 5.282 (3.739–7.461) | < 0.001 |
*ORs and 95% CIs calculated by logistic regression.
Adjusted OR(95%CI) and p value adjusted for age, body mass index, diabetes mellitus, hypertension, history of smoking, history of alcohol use, family history and hyperlipidemia.
OR, odds ratio; CI, confidence interval.
Table 10.
Stratified odds ratios for rs10739971 and rs17276588 with respect to the risk of ischemic stroke.
| Stratum | SNP | Male |
Female |
||
|---|---|---|---|---|---|
| OR (95%CI)*a | p valueb | OR (95%CI)*a | p valueb | ||
| Stratified by rs10739971 | |||||
| GA or AA | rs17276588 (AA vs GG) | 1.579 (1.056–2.361) | 0.026 | 2.373 (1.819–3.096) | < 0.001 |
| GG | rs17276588 (AA vs GG) | 2.656 (1.955–3.608) | < 0.001 | 6.813 (4.271–10.869) | < 0.001 |
| Stratified by rs17276588 | |||||
| GA or AA | rs10739971 (GA or AA vs GG) | 1.584 (1.017–2.469) | 0.042 | 0.683 (0.448–1.042) | 0.077 |
| GG | rs10739971 (AA vs GG) | 0.997 (0.786–1.264) | 0.979 | 1.705 (1.232–2.359) | < 0.001 |
*ORs and 95% CIs calculated by logistic regression.
Adjusted OR(95%CI) and p value adjusted for age, body mass index, diabetes mellitus, hypertension, history of smoking, history of alcohol use, family history and hyperlipidemia.
SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Discussion
The present study evaluated associations between two pri-miRNA SNPs (pri-let-7f-1 rs10739971 and pri-let-7f-2 rs17276588) and the risk of IS in a northern Chinese Han population. To the best of our knowledge, this is the first study to investigate these relationships in a large population (3259 subjects, 1803 patients and 1456 controls), as well as analyzing an X-linked locus, rs17276588, with the aim of exploring sex differences in IS risk. The findings indicated that both rs10739971 and rs17276588 polymorphisms were likely to be linked to an increased risk of IS. Individuals with the AA genotype of either SNP may have a higher risk of developing IS, with the A allele being a high-risk allele. The rs17276588 polymorphism tended to interact with a history of alcohol use, and alcohol use increased the risk of IS associated with rs17276588 mutation in women, indicating a synergistic effect. These results suggest that individuals with a high-risk genotype or allele, particularly women, should possibly avoid alcohol use. We also explored the interaction between the SNPs and demonstrated multiplicative and additive effects in both males and females. Certain groups of genotypes had combined effects, especially when several risk mutations occurred in the same individual. The lack of any GA genotype in males led to a slight difference in rs17276588, but the risk allele remained unchanged.
Let-7 family members have been widely studied as tumor suppressors. Let-7, as a ligand, has been reported to curb glioma growth via Toll-like receptor 7 in the brain, and was associated with microglial function. 28 The let-7 family has also been investigated in relation to various neurological diseases and has been correlated with neurodegenerative disorders 29 , while let-7b-5p levels in cerebrospinal fluid were reduced in patients with progressive multiple sclerosis. 30 Let-7b may be involved in the regulation of hepatic stellate cells 31 , and let-7 family members have also been shown to suppress inflammation.32–34 Our previous study on let-7a suggested a relationship between the rs1143770 and rs629367 SNPs on chromosome 11 and IS, 24 thus laying the foundations for the current study.
As a let-7 family member, let-7f plays a key role in many pathological processes during tumor development, including cell migration 35 and differentiation. 36 It was also shown to be involved in endothelial function and angiogenesis, 37 and a let-7f antagomir showed a neuroprotective effect. 38 Let-7f expression increased gradually during brain development and was shown to promote the differentiation of neural stem cells in rat models, 39 establishing a potential association between let-7f and brain disease. Another study showed that inhibition of let-7f increased the expression of the antiangiogenic protein, thrombospondin-1, 40 suggesting that increased expression of let-7f in the brain increased the risk of thrombosis. An antagomir to let-7f provided neuroprotection in an ischemic model by inhibiting let-7f expression and boosting the endogenous neuroprotective agent, insulin-like growth factor-1, which plays an important role in the evolution of acute cerebral ischemia. 41 In addition, anti-let-7f oligonucleotides diminished cortical and striatal infarcts and preserved sensorimotor function and interhemispheric neural integration. 38 The endothelial function of let-7f was analyzed in young stroke patients, and it was considered to act as a biomarker in the diagnosis and prognosis of cerebral IS. 42
The fact that pri-miRNAs affect miRNA expression via Drosha, the catalytic subunit of the microprocessor complex, suggests a possible underlying mechanism. 43 Previous studies of pri-let-7f rs10739971 and rs17276588 SNPs mostly focused on distinct types of cancer: Yuan et al. found that decreased transcription of rs17276588 A allele led to reduced levels of let-7f and a higher risk of colorectal cancer, 23 while pri-let-7f-1 rs10739971 polymorphism together with ERCC6 and PGC polymorphisms served as a predictive model for the risk of gastric cancer. 44 Few studies have investigated the direct roles of these two SNPs in IS; however, the GA and AA variant genotypes of rs17276588 were associated with an enhanced risk of metabolic syndrome, 45 which is a multi-component disease caused by abnormal metabolic system abnormalities, leading to multiple clinical disorders including diabetes and dyslipidemia. 46 The influence of rs17276588 SNPs on metabolism not only sheds light on the prospective link between SNPs and IS, but also helps to explain the sex-related differences in its effects, via hormonal effects.
In this study, we assumed that rs10739971 and rs17276588 polymorphisms were associated with IS risk and tested this hypothesis in a hospital-based case–control study. We carried out statistical analysis using dominant and recessive models to estimate the mutations and their corresponding traits, and examined gene–environment and gene–gene interactions, including both multiplicative and additive effects, to determine any extra impact on IS risk. However, it was not possible to reach a definite conclusion, despite the relatively large population involved in this study. This study had some limitations. First, we focused on a northern Chinese Han population, and more data are needed, especially from different regions and races. Second, this study failed to determine the specific gene–disease mechanism. Third, we did not carry out in vivo and in vitro experiments to test the results. However, despite these shortcomings, the study also had several strengths, and the comprehensive analyses help to uncover the underlying relationship.
In conclusion, the results of the current study suggest that the AA genotypes and A alleles of pri-let-7f-1 rs10739971 and pri-let-7f-2 rs17276588 are associated with an increased risk of IS. There is also a gene–environment interaction and a slight sex difference in the effect of rs17276588, suggesting that women with the A allele should play close attention to their alcohol consumption. These results support the use of these two SNPs as valuable biomarkers for caution, and individuals with high-risk alleles should undergo regular examinations and be careful regarding other conventional risk factors, to reduce the occurrence of IS. Further studies with larger sample sizes are needed to confirm these results and to investigate the potential mechanisms underlying the relationship between let-7f and IS.
Research data
Research Data for Genetic polymorphisms of pri-let-7f, gene–environment and gene–gene interactions, and associations with ischemic stroke risk in Liaoning Province by Luying Qiu, Yuye Wang, Fang Liu, Shumin Deng, Zhiyi He, Wenxu Zheng and Yanzhe Wang in Journal of International Medical Research
Footnotes
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the National Natural Science Foundation of China [grant number 81901189], and by the Natural Science Foundation of Liaoning Province of China [grant number 2019-BS-147] (both to YZW). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication
ORCID iDs: Luying Qiu https://orcid.org/0000-0003-0850-3393
Availability of data and materials
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Authors’ contributions
LYQ carried out this study and drafted the manuscript. YYW prepared the figures and interpreted the data. SMD and FL conceived the project. ZYH, WXZ, and YZW revised and finalized the manuscript. All authors read and approved the final manuscript.
Declaration of conflicting interests
The authors declare that there is no conflict of interest.
References
- 1.Donkor ES.Stroke in the 21(st) century: A snapshot of the burden, epidemiology, and quality of life. Stroke Res Treat 2018; 2018: 3238165–3238165. DOI: 10.1155/2018/3238165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GBD 2016 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017; 390: 1260–1344. 2017/09/19. DOI: 10.1016/s0140-6736(17)32130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feigin VL, Nguyen G, Cercy K, et al. Global, regional, and country-specific lifetime risks of stroke, 1990 and 2016. N Engl J Med 2018; 379: 2429–2437. 2018/12/24. DOI: 10.1056/NEJMoa1804492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tu WJ, Hua Y, Yan F, et al. Prevalence of stroke in China, 2013-2019: A population-based study. Lancet Reg Health West Pac 2022; 28: 100550. 2022/12/13. DOI: 10.1016/j.lanwpc.2022.100550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bamford J, Sandercock P, Dennis M, et al. Classification and natural history of clinically identifiable subtypes of cerebral infarction. Lancet 1991; 337: 1521–1526. 1991/06/22. DOI: 10.1016/0140-6736(91)93206-o. [DOI] [PubMed] [Google Scholar]
- 6.O'Donnell MJ, Xavier D, Liu L, et al. Risk factors for ischaemic and intracerebral haemorrhagic stroke in 22 countries (the INTERSTROKE study): a case-control study. Lancet 2010; 376: 112–123. 2010/06/22. DOI: 10.1016/s0140-6736(10)60834-3. [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez C, Ramos-Araque ME, Domínguez-Martínez M, et al. Single-nucleotide polymorphism 309T>G in the MDM2 promoter determines functional outcome after stroke. Stroke 2018; 49: 2437–2444. 2018/10/26. DOI: 10.1161/strokeaha.118.022529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talebi M, Delpak A, Khalaj-Kondori M, et al. ABCA7 and EphA1 genes polymorphisms in late-onset Alzheimer’s disease. J Mol Neurosci 2020; 70: 167–173. 2019/10/30. DOI: 10.1007/s12031-019-01420-x. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP.MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. 2004/01/28. DOI: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Zhu R, Zhao Y, Xiao T, et al. Association between microRNA binding site polymorphisms in immunoinflammatory genes and recurrence risk of ischemic stroke. Genomics 2020; 112: 2241–2246. 2019/12/29. DOI: 10.1016/j.ygeno.2019.12.020. [DOI] [PubMed] [Google Scholar]
- 11.Matsuyama H, Suzuki HI.Systems and synthetic microRNA biology: from biogenesis to disease pathogenesis. Int J Mol Sci 2019; 21: 132. 2019/12/28. DOI: 10.3390/ijms21010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandolfini L, Barbieri I, Bannister AJ, et al. METTL1 promotes let-7 microRNA processing via m7G methylation. Mol Cell 2019; 74: 1278–1290.e9. 2019/04/30. DOI: 10.1016/j.molcel.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roush S, Slack FJ.The let-7 family of microRNAs. Trends Cell Biol 2008; 18: 505–516. 2008/09/09. DOI: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005; 353: 1793–1801. 2005/10/28. DOI: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 15.Gill D, James NE, Monori G, et al. Genetically determined risk of depression and functional outcome after ischemic stroke. Stroke 2019; 50: 2219–2222. 2019/06/27. DOI: 10.1161/strokeaha.119.026089. [DOI] [PubMed] [Google Scholar]
- 16.Fang S, Hu X, Wang T, et al. Parkinson's disease and ischemic stroke: a bidirectional Mendelian randomization study. Transl Stroke Res 2022; 13: 528–532. 2022/01/12. DOI: 10.1007/s12975-021-00974-6. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Yin X, Li L, et al. Association of apolipoprotein C3 genetic polymorphisms with the risk of ischemic stroke in the Northern Chinese Han population. PloS One 2016; 11: e0163910. 2016/10/01. DOI: 10.1371/journal.pone.0163910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Liu F, Li L, et al. The association between apolipoprotein A1-C3-A5 gene cluster promoter polymorphisms and risk of ischemic stroke in the northern Chinese Han population. The J Int Med Res 2017; 45: 2042–2052. 2017/06/22. DOI: 10.1177/0300060517713517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang YZ, Zhang HY, Liu F, et al. Association between PPARG genetic polymorphisms and ischemic stroke risk in a northern Chinese Han population: a case-control study. Neural Regen Res 2019; 14: 1986–1993. 2019/07/11. DOI: 10.4103/1673-5374.259621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Xu Q, Liu J, et al. Pri-let-7a-1 rs10739971 polymorphism is associated with gastric cancer prognosis and might affect mature let-7a expression. Onco Targets Ther 2016; 9: 3951–3962. 2016/07/23. DOI: 10.2147/ott.S100481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin KM, Jung DK, Hong MJ, et al. The pri-let-7a-2 rs1143770C>T is associated with prognosis of surgically resected non-small cell lung cancer. Gene 2016; 577: 148–152. 2015/12/03. DOI: 10.1016/j.gene.2015.11.036. [DOI] [PubMed] [Google Scholar]
- 22.Yao Y, Wang W, Jing L, et al. Let-7f regulates the hypoxic response in cerebral ischemia by targeting NDRG3. Neurochem Res 2017; 42: 446–454. DOI: 10.1007/s11064-016-2091-x. [DOI] [PubMed] [Google Scholar]
- 23.Yuan F, Xiao X, Che G, et al. A functional variant in the flanking region of pri-let-7f contributes to colorectal cancer risk in a Chinese population. J Cell Physiol 2019; 234: 15717–15725. 2019/02/12. DOI: 10.1002/jcp.28227. [DOI] [PubMed] [Google Scholar]
- 24.Wang YY, Zhang HY, Jiang WJ, et al. Genetic polymorphisms in pri-let-7a-2 are associated with ischemic stroke risk in a Chinese Han population from Liaoning, China: a case-control study. Neural Regen Res 2021; 16: 1302–1307. 2020/12/16. DOI: 10.4103/1673-5374.301019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssens AC, Ioannidis JP, Van Duijn CM, et al. Strengthening the reporting of Genetic RIsk Prediction Studies: the GRIPS Statement. PLoS Med 2011; 8: e1000420. 2011/03/23. DOI: 10.1371/journal.pmed.1000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leng RX, Pan HF, Liu J, et al. Evidence for genetic association of TBX21 and IFNG with systemic lupus erythematosus in a Chinese Han population. Sci Rep 2016; 6: 22081. 2016/02/27. DOI: 10.1038/srep22081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersson T, Alfredsson L, Källberg H, et al. Calculating measures of biological interaction. Eur J Epidemiol 2005; 20: 575–579. 2005/08/27. DOI: 10.1007/s10654-005-7835-x. [DOI] [PubMed] [Google Scholar]
- 28.Buonfiglioli A, Efe IE, Guneykaya D, et al. Let-7 microRNAs regulate microglial function and suppress glioma growth through Toll-like receptor 7. Cell Rep 2019; 29: 3460–3471.e7. 2019/12/12. DOI: 10.1016/j.celrep.2019.11.029. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Qi Y, Liu CF, et al. MicroRNA expression data analysis to identify key miRNAs associated with Alzheimer’s disease. J Gene Med 2018; 20: e3014. 2018/03/16. DOI: 10.1002/jgm.3014. [DOI] [PubMed] [Google Scholar]
- 30.Mandolesi G, Rizzo FR, Balletta S, et al. The microRNA let-7b-5p is negatively associated with inflammation and disease severity in multiple sclerosis. Cells 2021; 10: 330. 2021/02/11. DOI: 10.3390/cells10020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun X, Zhang L, Jiang Y, et al. The role of let-7b in the inhibition of hepatic stellate cell activation by rSjP40. PLoS Negl Trop Dis 2021; 15: e0009472. 2021/06/24. DOI: 10.1371/journal.pntd.0009472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rom S, Dykstra H, Zuluaga-Ramirez V, et al. miR-98 and let-7g* protect the blood-brain barrier under neuroinflammatory conditions. Cereb Blood Flow Metab 2015; 35: 1957–1965. 2015/07/02. DOI: 10.1038/jcbfm.2015.154.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jickling GC, Ander BP, Shroff N, et al. Leukocyte response is regulated by microRNA let7i in patients with acute ischemic stroke. Neurology 2016; 87: 2198–2205. 2016/10/28. DOI: 10.1212/wnl.0000000000003354.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan H, Zhang H, Hong L, et al. MicroRNA let-7c-5p suppressed lipopolysaccharide-induced dental pulp inflammation by inhibiting dentin matrix protein-1-mediated nuclear factor kappa B (NF-κB) pathway in vitro and in vivo. Med Science Monit 2018; 24: 6656–6665. 2018/09/22. DOI: 10.12659/msm.909093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue H, Gao X, Xu S, et al. MicroRNA-Let-7f reduces the vasculogenic mimicry of human glioma cells by regulating periostin-dependent migration. Oncol Rep 2016; 35: 1771–1777. 2016/01/12. DOI: 10.3892/or.2016.4548. [DOI] [PubMed] [Google Scholar]
- 36.Rogucki M, Buczyńska A, Krętowski AJ, et al. The importance of miRNA in the diagnosis and prognosis of papillary thyroid cancer. J Clin Med 2021; 10: 4738. 2021/10/24. DOI: 10.3390/jcm10204738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urbich C, Kuehbacher A, Dimmeler S.Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res 2008; 79: 581–588. 2008/06/14. DOI: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 38.Selvamani A, Sathyan P, Miranda RC, et al. An antagomir to microRNA Let7f promotes neuroprotection in an ischemic stroke model. PloS One 2012; 7: e32662. 2012/03/07. DOI: 10.1371/journal.pone.0032662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng Z, Wei Y, Yao Y, et al. Let-7f promotes the differentiation of neural stem cells in rats. Am J Transl Res 2020; 12: 5752–5761. 2020/10/13. [PMC free article] [PubMed] [Google Scholar]
- 40.Tao WY, Liang XS, Liu Y, et al. Decrease of let-7f in low-dose metronomic paclitaxel chemotherapy contributed to upregulation of thrombospondin-1 in breast cancer. Int J Biol Sci 2015; 11: 48–58. 2015/01/02. DOI: 10.7150/ijbs.9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwab S, Spranger M, Krempien S, et al. Plasma insulin-like growth factor I and IGF binding protein 3 levels in patients with acute cerebral ischemic injury. Stroke 1997; 28: 1744–1748. 1997/09/26. DOI: 10.1161/01.str.28.9.1744. [DOI] [PubMed] [Google Scholar]
- 42.Tan KS, Armugam A, Sepramaniam S, et al. Expression profile of MicroRNAs in young stroke patients. PloS One 2009; 4: e7689. 2009/11/06. DOI: 10.1371/journal.pone.0007689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin W, Wang J, Liu CP, et al. Structural basis for pri-miRNA recognition by Drosha. Mol Cell 2020; 78: 423–433.e5. 2020/03/30. DOI: 10.1016/j.molcel.2020.02.024. [DOI] [PubMed] [Google Scholar]
- 44.Xu Q, Liu JW, He CY, et al. The interaction effects of pri-let-7a-1 rs10739971 with PGC and ERCC6 gene polymorphisms in gastric cancer and atrophic gastritis. PloS One 2014; 9: e89203. 2014/03/04. DOI: 10.1371/journal.pone.0089203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan YX, Wu LJ, Zhang J, et al. Let-7 related genetic variation and risk of metabolic syndrome in a Chinese population. Endocr J 2015; 62: 887–896. 2015/07/17. DOI: 10.1507/endocrj.EJ15-0236. [DOI] [PubMed] [Google Scholar]
- 46.Moller DE, Kaufman KD.Metabolic syndrome: a clinical and molecular perspective. Annu Rev Med 2005; 56: 45–62. 2005/01/22. DOI: 10.1146/annurev.med.56.082103.104751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Research Data for Genetic polymorphisms of pri-let-7f, gene–environment and gene–gene interactions, and associations with ischemic stroke risk in Liaoning Province by Luying Qiu, Yuye Wang, Fang Liu, Shumin Deng, Zhiyi He, Wenxu Zheng and Yanzhe Wang in Journal of International Medical Research
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.