Figure 1. Transposon mutagenesis reveals the essential gene set of a chemolithoautotrophic organism.
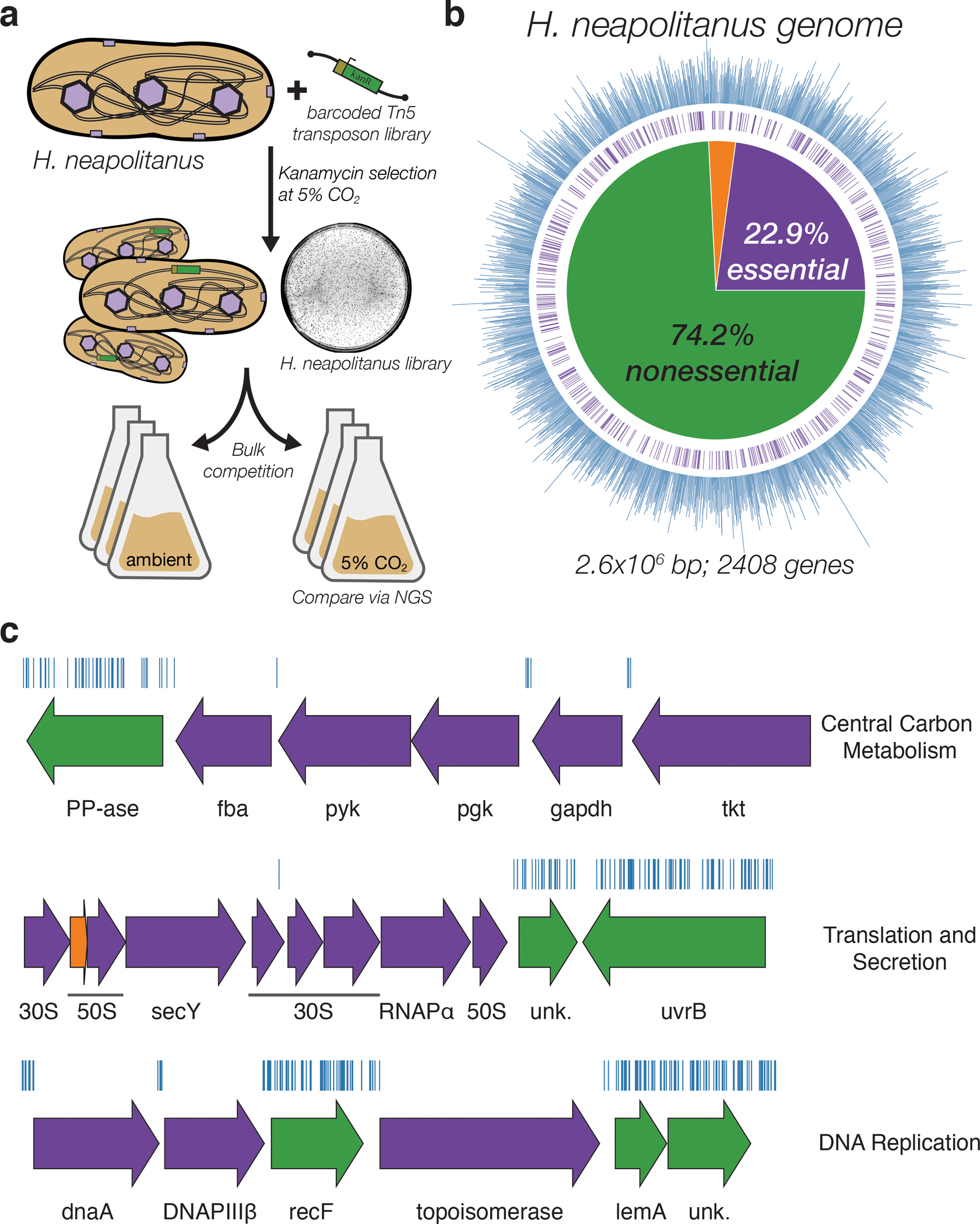
a. Schematic depicting the generation and screening of the RB-TnSeq library. Transposons were inserted into the Hnea genome by conjugation with an E. coli donor strain. The transposon contains a random 20 base pair barcode (yellow) and a kanamycin selection marker (green). Selection for colonies containing insertions was performed in the presence of kanamycin at 5% CO2 and insertions were mapped by sequencing as described in the Methods. Subsequent screens were carried out as bulk competition assays and quantified by BarSeq. b. Insertions and essential genes are well-distributed throughout the Hnea genome. The outer track (blue) is a histogram of the number of barcodes that were mapped to a 1 kb window. The inner track annotates essential genes in purple. The pie chart shows the percentages of the genome called essential (purple), ambiguous (orange), and nonessential (green). c. Representative essential genes and nonessential genes in the Hnea genome. The blue track indicates the presence of an insertion. Genes in purple were called essential and genes in green are nonessential. Genes labeled “unk.” are hypothetical proteins. The first genomic locus contains 5 essential genes involved in glycolysis or the CBB cycle including pyruvate kinase (pyk) and transketolase (tkt). The 8 essential genes in the second locus encoding 30S and 50S subunits of the ribosome, the secY secretory channel, and an RNA polymerase subunit. Essential genes in the third example locus include topoisomerase and DNA polymerase III β. A full analysis with gene names is in Supplemental Figure 1 and essentiality information for every gene can be found in supplemental file 2.
