Figure 2. A systematic screen for high CO2-requiring mutants identifies genes putatively associated with the CCM.
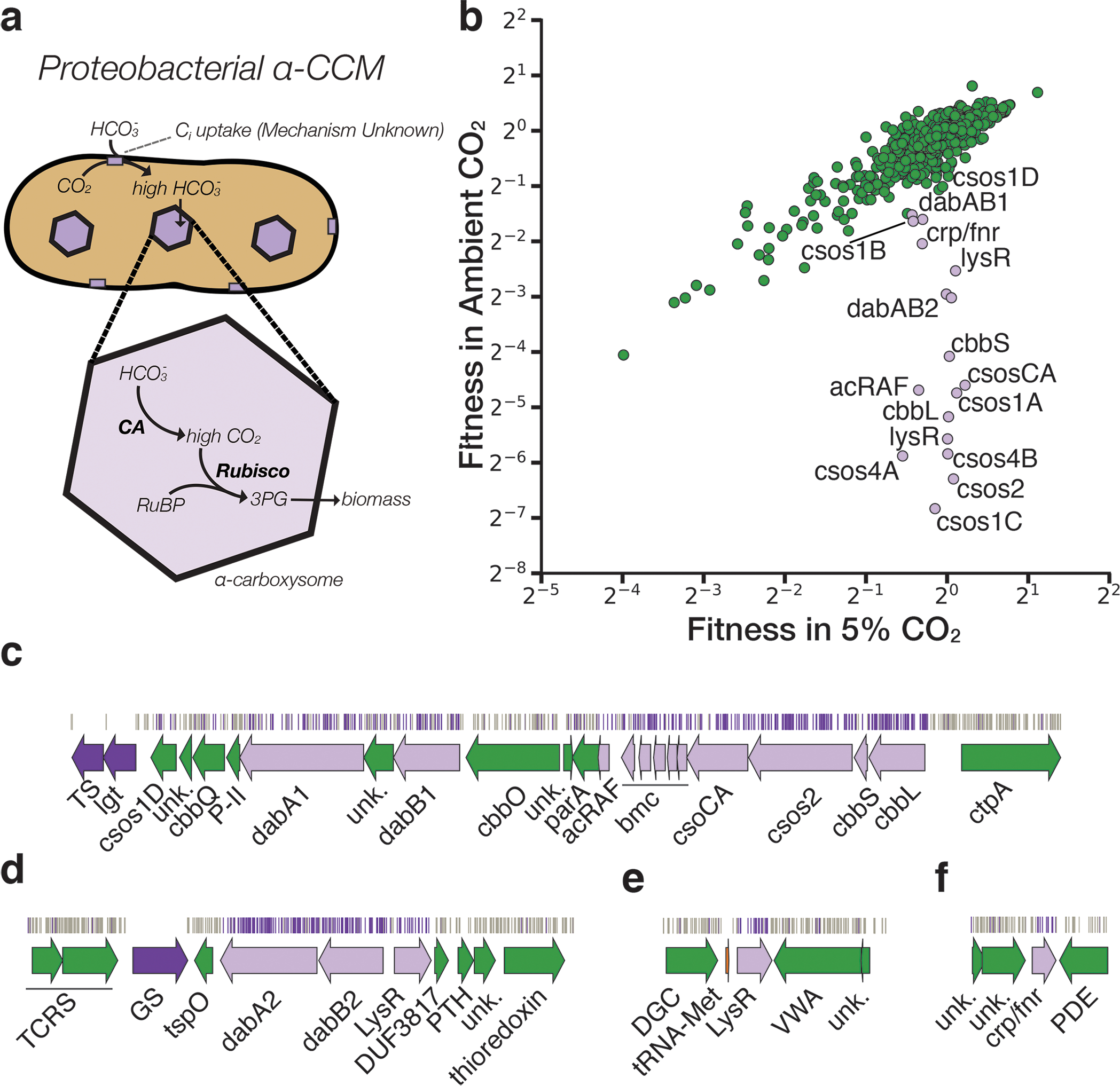
a. Simplified model of the ɑ-CCM of chemolithoautotrophic proteobacteria. Inorganic carbon is concentrated via an unknown mechanism, producing a high cytosolic HCO3− concentration. High cytosolic HCO3− is converted into high carboxysomal CO2 by CA, which is localized only to the carboxysome. b. Fitness effects of gene knockouts in 5% CO2 as compared to ambient CO2. Data is from one of two replicates of BarSeq. The effects of single transposon insertions into a gene are averaged to produce the gene-level fitness value plotted. We define HCR mutants as those displaying a twofold fitness defect in ambient CO2 relative to 5% CO2 in both replicates. HCR genes are colored light purple. Data from both replicates and the associated standard errors are shown in Supplemental Figure 2 and in supplemental file 3. Panels c-f show regions of the Hnea genome containing genes annotated as HCR in panel A. Essential genes are in dark purple, HCR genes are in light purple, and other genes are in green. The top tracks show the presence of an insertion in that location. Insertions are colored grey unless they display a twofold or greater fitness defect in ambient CO2, in which case they are colored light purple. c. The gene cluster containing the carboxysome operon and a second CCM-associated operon. This second operon contains acRAF, a Form IC associated cbbOQ-type Rubisco activase and dabAB1. d. The DAB2 operon and surrounding genomic context. e. The genomic context of a lysR-type transcriptional regulator that shows an HCR phenotype. f Genomic context of a crp/fnr-type transcriptional regulator that displays an HCR phenotype. Genes labeled “unk.” are hypothetical proteins. Full gene names are given in Supplemental Figure 3. Accession numbers and gi numbers for selected genes can be found in Supplemental Table 1.
