Figure 3. The DABs catalyze active transport of Ci and are energized by a cation gradient.
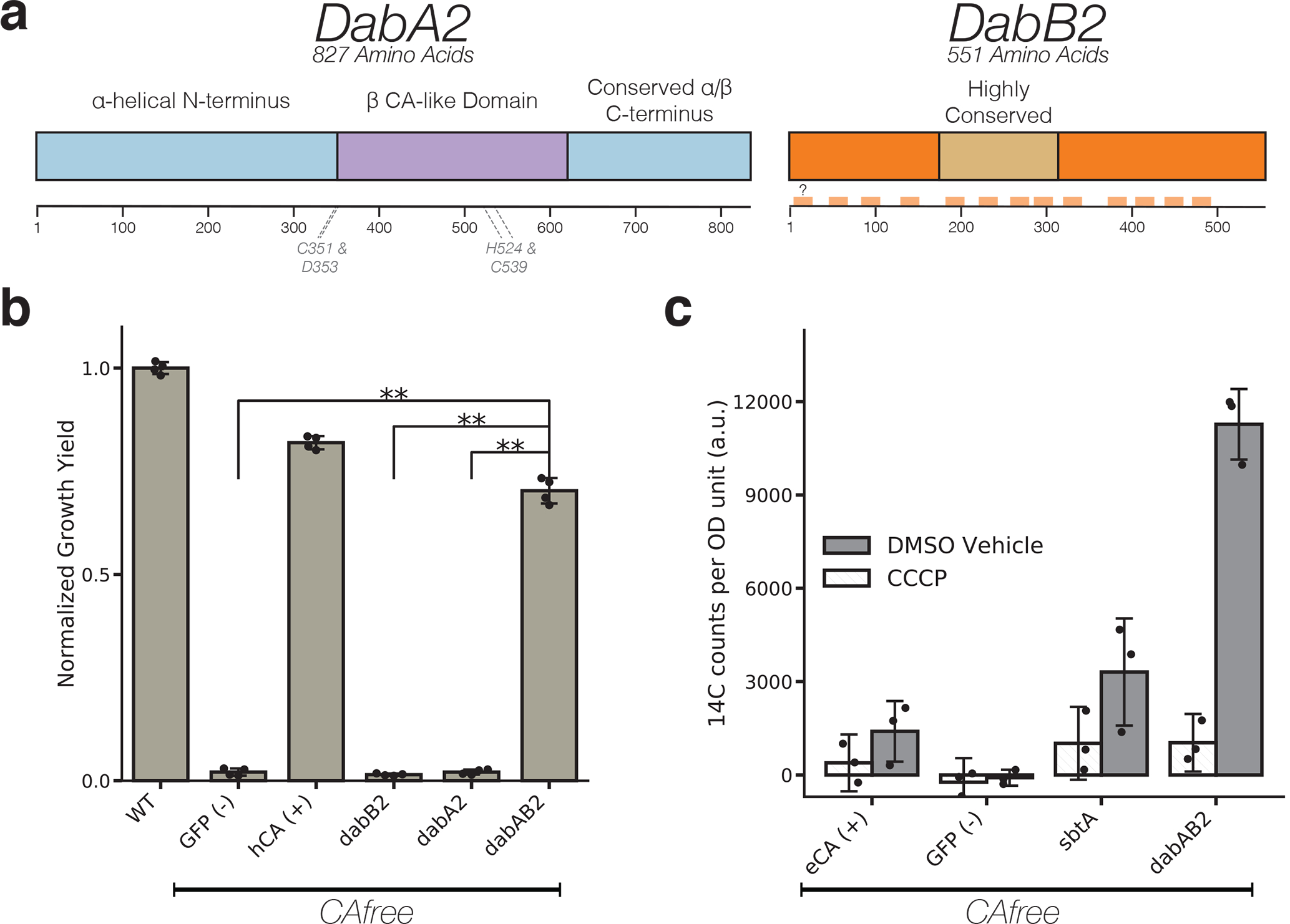
a. Diagrammatic representation of DabA2 and DabB2 based on bioinformatic annotation. The four predicted active site residues (C351, D353, H524, C539) are marked on the primary amino acid sequence. Amino acid numbers are marked below each gene and predicted transmembrane helices are marked in light orange. b. DAB2 was tested for ability to rescue growth of CAfree E. coli in ambient CO2 conditions. Expression of the full operon (DabAB2) rescues growth, as does the positive control, and human carbonic anhydrase II (hCA). dabAB2 has a larger rescue than GFP (t=42.6, corrected p=3.4*10−8), dabA2 (t=43.4, corrected p=3*10−8), and dabB2 (t=44.5, corrected p=2.6*10−8). “**” denotes that means are significantly different with Bonferroni corrected p < 5X10−4 according to a two-tailed t-test. Bar heights represent means and error bars represent standard deviations of 4 biological replicates. Consistent results were seen after 2 independent experiments. c. CAfree E. coli were tested for Ci uptake using the silicone-oil centrifugation method. Expression of DabAB2 produced a large increase in 14C uptake as compared to all controls. Moreover, treatment with the ionophore CCCP greatly reduces DabAB2-mediated 14C uptake, suggesting that DabAB2 is coupled to a cation gradient. E. coli CA (eCA) was used as a control for a non-vectorial CA. Synechococcus elongatus PCC 7942 sbtA was used as a known Ci transporter. GFP was used as a vector control. Bar heights represent the mean and error bars represent standard deviations of 3 technical replicates. Consistent results were seen with 3 independent experiments.
