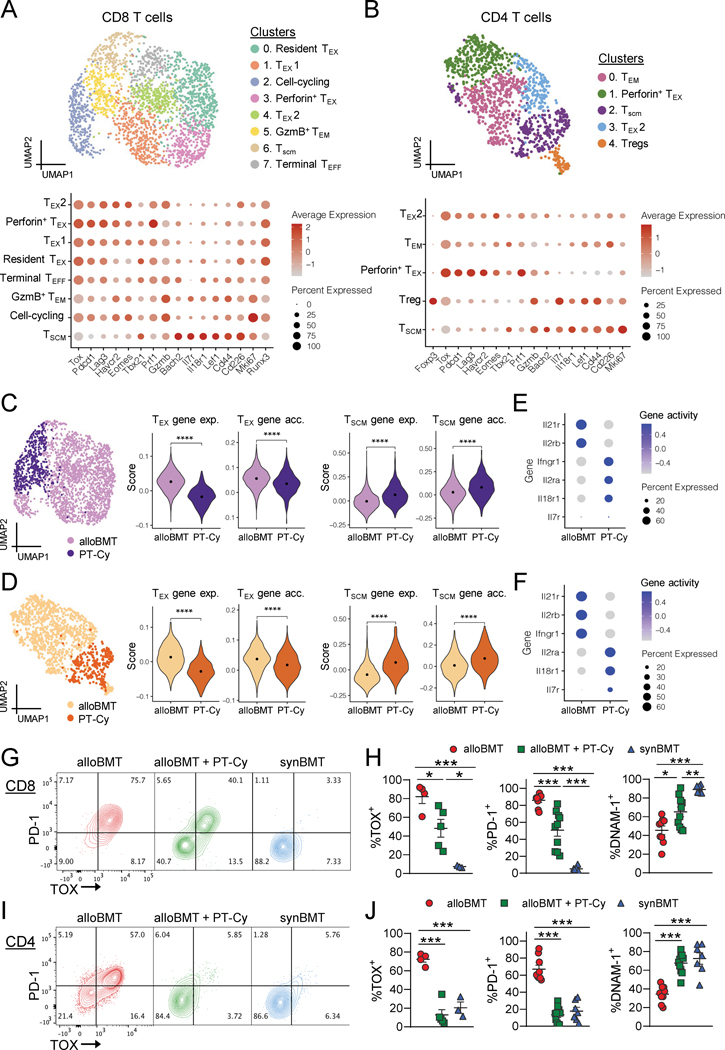
Figure 4: PT-Cy reduced alloreactive T cell exhaustion and enhanced stemness in bone marrow.
B6 recipients were transplanted with 5 × 106 BM with 0.5 × 106 CD4+ + 0.5 × 106 CD8+ T cells from C3H.SW donors (alloBMT) or B6 donors (synBMT). Some alloBMT recipients were treated with 50 mg/kg cyclophosphamide on D+3 and D+4 after transplantation (alloBMT + PTCy). Mice were sacrificed 14 days after transplant and BM was harvested and pooled from 4 mice per group. (A-F) CD8+ and CD4+ T cells were sort purified from BM of alloBMT and alloBMT + PT-Cy recipients and nuclei were processed for 10x genomics multiome sequencing. (A) WNN embedding of combined ATAC and RNA data of CD8+ cells colored by cluster (top) than annotated using CD8+ T cell specific markers (bottom). (B) CD4+ T cells clustered and annotated in a manner analogous to (A). (C) Embedding in (A) colored by experimental group (left). Centered and scaled cumulative gene expression (abbrev. ‘exp’) and gene accessibility (abbrev. ‘acc’, using gene activity score) of TEX and TSCM genes in CD8+ T cells by experimental group (right). Wilcoxon Rank Sum test. (D) Embedding in (B) colored by experimental group (left). Centered and scaled cumulative gene expression (abbrev. ‘exp’) and gene accessibility of TEX and TSCM genes in CD4+ T cells by experimental group (right). Wilcoxon Rank Sum test. (E-F) Gene accessibility scores of key cytokine receptor genes by experiment group in (E) CD8+ T cells and (F) CD4+ T cells. (G-J) Representative flow cytometry plots of PD-1 versus TOX expression in CD8+ and CD4+ conventional (FoxP3−) T cells. (H) Frequency of TOX+, PD-1+ and DNAM-1+ within CD8+ T cells (J) and CD4+ conventional T cells. (n = 7–10 /group from 2 experiments, TOX n = 3 – 5 /group from 1 experiment). Data represent mean ± SEM. One-way ANOVA with Tukey’s multiple comparisons test. * p<0.05, ** p<0.01, ***p<0.001, ****p<0.0001.