Figure 2. DedA and DUF368 family members provide resistance to UndP-targeting antibiotics.
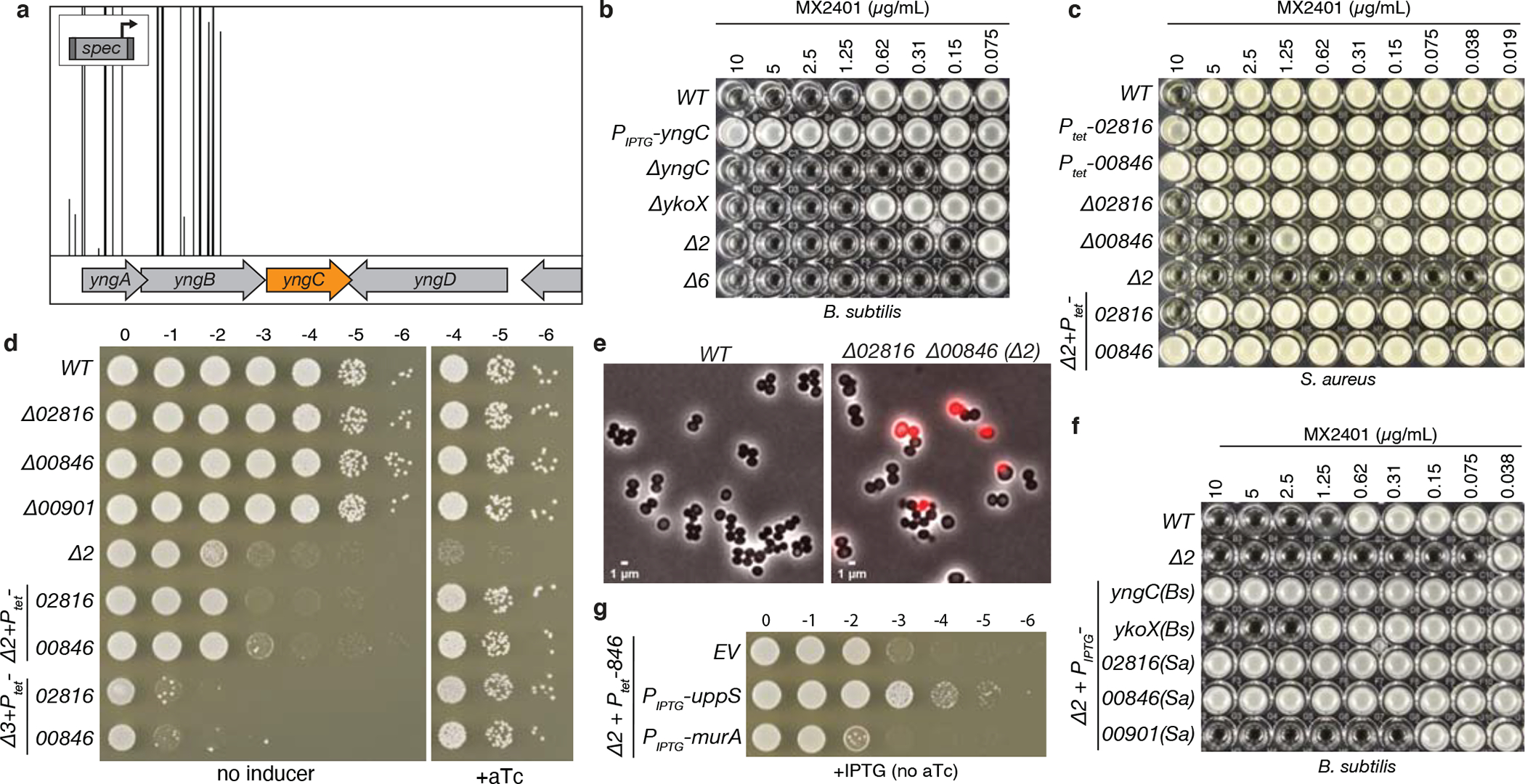
(a) A transposon-sequencing screen for MX2401-resistance mutants identifies yngC. Transposon insertion profile at the indicated B. subtilis genomic region is shown. Each vertical line indicates a Tn insertion site; its height reflects the number of sequencing reads at this position. All transposons in this region were inserted with the outward facing promoter facing yngC as indicated in the schematic. Most insertion sites had >20,000 reads but the window size was scaled to 5,000 to highlight the absence of reads in the neighboring genes. (b) Minimum inhibitory concentration (MIC) assay of the indicated B. subtilis strains. (c) MIC assay of the indicated S. aureus strains. (d) Spot-dilution assays of the indicated S. aureus strains on TSA with or without 100 ng/mL anhydrotetracycline (+aTc). (e) Representative images of wild-type and Δ00846 Δ02816 (Δ2) S. aureus cells. Overlays of phase-contrast and propidium iodide fluorescence are shown. (f) MIC assays of the indicated B. subtilis strains over-expressing DedA and DUF368 family members from B. subtilis and S. aureus. (g) Spot-dilutions of the indicated S. aureus strains harboring an empty vector (EV) or a vector with an IPTG-regulated promoter fused to uppS or murA.
