Extended Data Fig. 5. Gene neighborhood analyses reveal that dedA paralogs can be found adjacent to or fused with pap2 lipid phosphatases or adjacent to uppP undecaprenyl-pyrophosphate phosphatases.
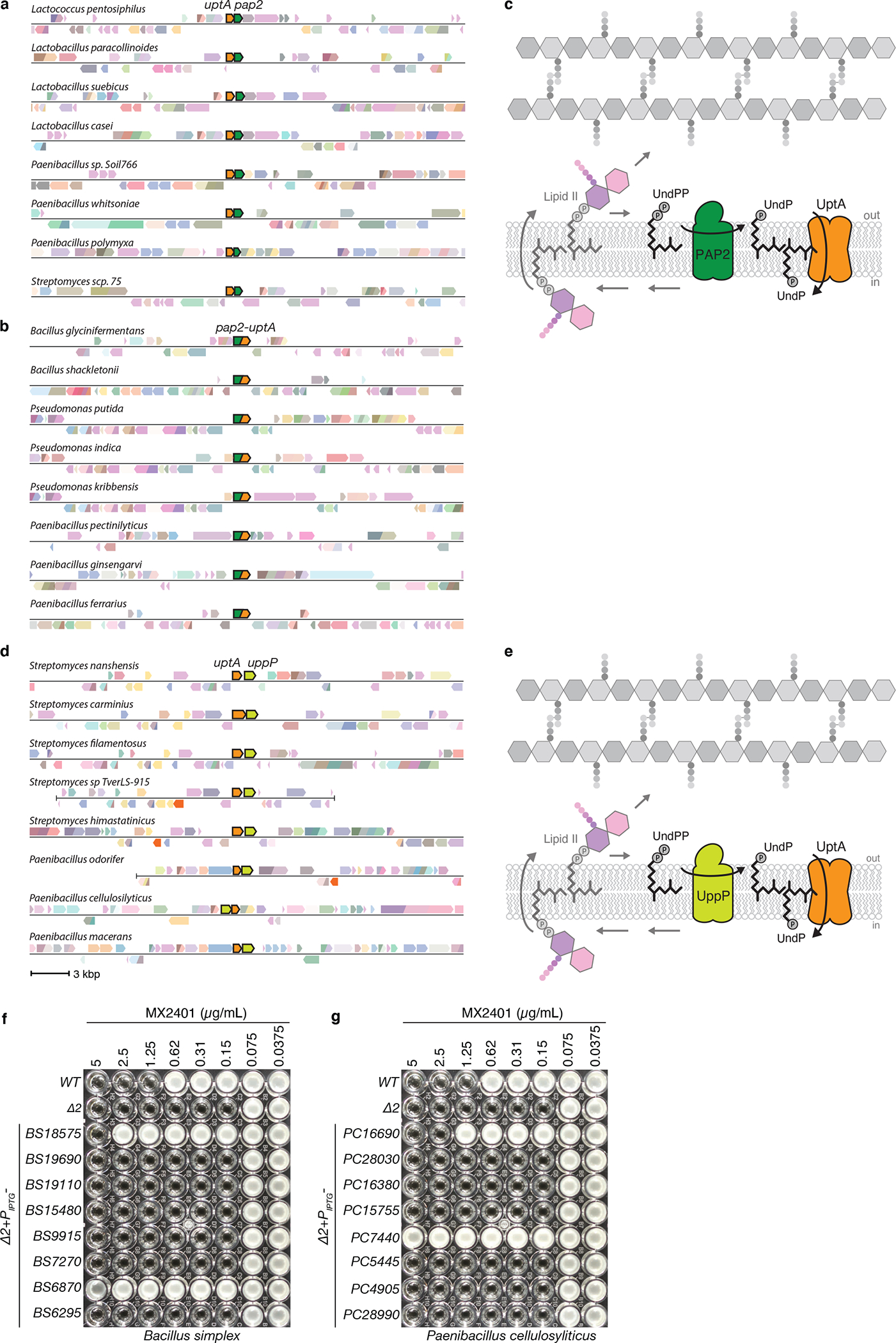
Representative genomic neighborhood analyses showing synteny (a) and gene-fusions (b) of DedA family members with PAP2 lipid phosphatases. (c) Schematic of the lipid II cycle highlighting the role of PAP2 family members like BcrC and DedA family members like YngC (UptA) in dephosphorylating UndPP and flipping UndP across the membrane, respectively. (d) Representative genomic neighborhood analysis showing synteny of dedA transporters with the undecaprenyl-pyrophosphate phosphatase uppP. Most examples of synteny between dedA and uppP genes are from Streptomycetes and Paenibacilli genomes. (e) Schematic of the lipid II cycle highlighting the role of UppP family members and DedA family members like YngC (UptA) in dephosphorylating UndPP and flipping UndP across the membrane, respectively. MIC assays of the indicated B. subtilis strains lacking uptA and ykoX (Δ2) expressing one of the eight Bacillus simplex (f) or Paenibacillus cellulosyliticus (g) DedA paralogs. BS18575 is adjacent to a pap2 gene in B. simplex. PC16690 is adjacent to a uppP gene in P. cellulosyliticus. Strains were grown in LB with 500 μM IPTG in the presence of the indicated concentration of MX2401. Uniprot IDs for the proteins included in the genome neighborhood diagrams can be found in Supplementary Table 3.
