Fig. 3. Prediction of conservation status of species using genomic information.
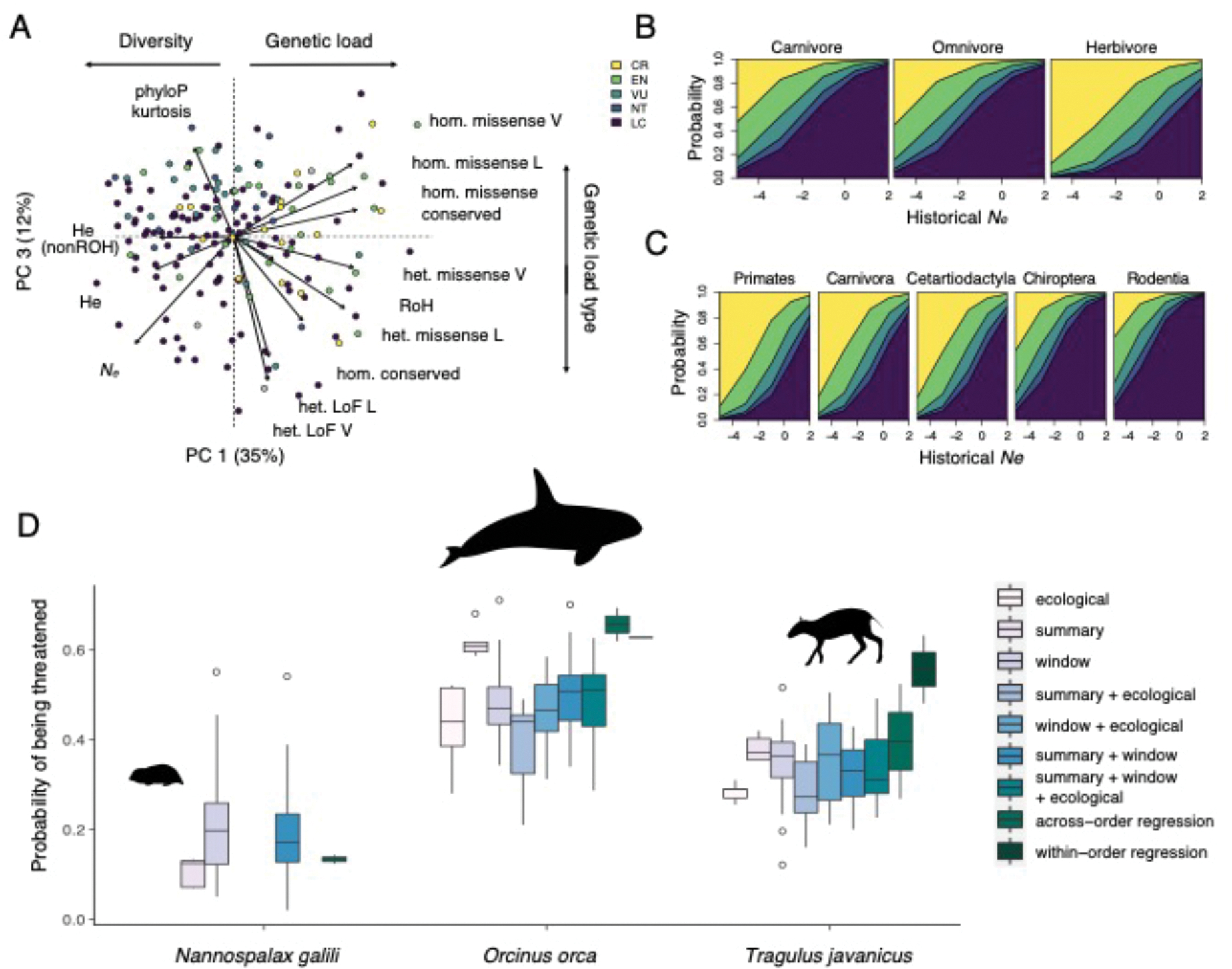
(A) Principal components (PCs) that significantly predict threatened status. PC1 describes heterozygosity, Ne and deleterious variation, and PC3 distinguishes types of deleterious variation. Loadings of genomic variables (arrows; table S3) are labeled as described in table S2 (L=IMPC lethal genes; V=IMPC viable genes). Points indicate species, colored by IUCN status as shown in (B). (B-C) Probability of assignment to IUCN categories by diet and scaled values of historical Ne (B), and by taxonomic order and historical Ne of species (C). Decreased historical Ne is consistently associated with increased risk, but the magnitude varies by diet and taxonomic order. (D) Conservation status predictions for three data deficient species using random forest models with window-based metrics (windows), ecological variables (ecological), and/or genome-wide summary variables (summary), and predictions from regression models within and across taxonomic orders. Nannospalax galili lacked ecological data and adequate within-order data, so only predictions from across-order regression and windows models are shown for this species.
