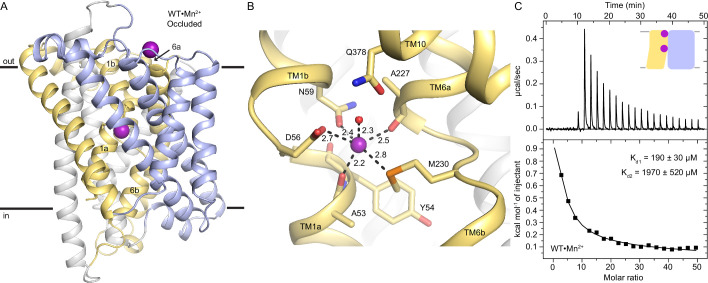
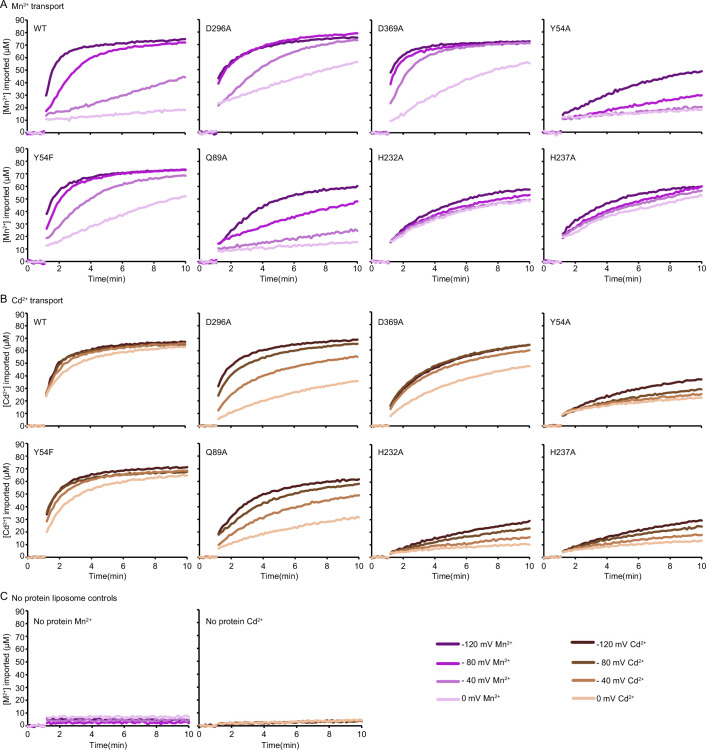
Figure 1. The occluded structure of DraNramp reveals a largely dehydrated Mn2+-coordination sphere.
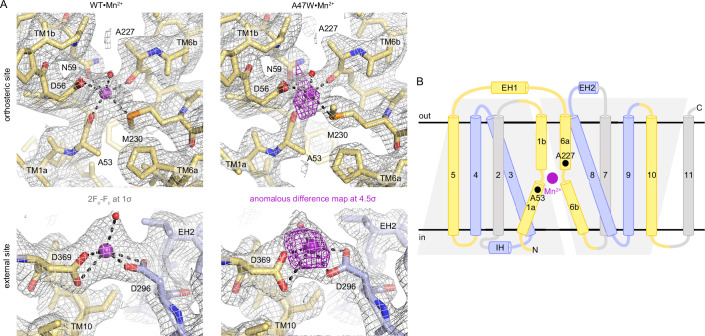
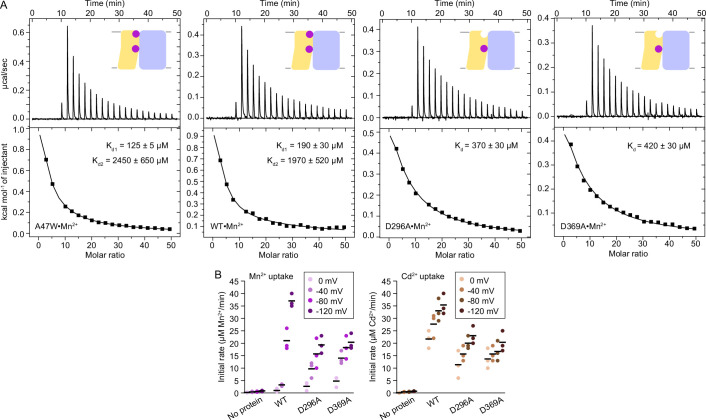
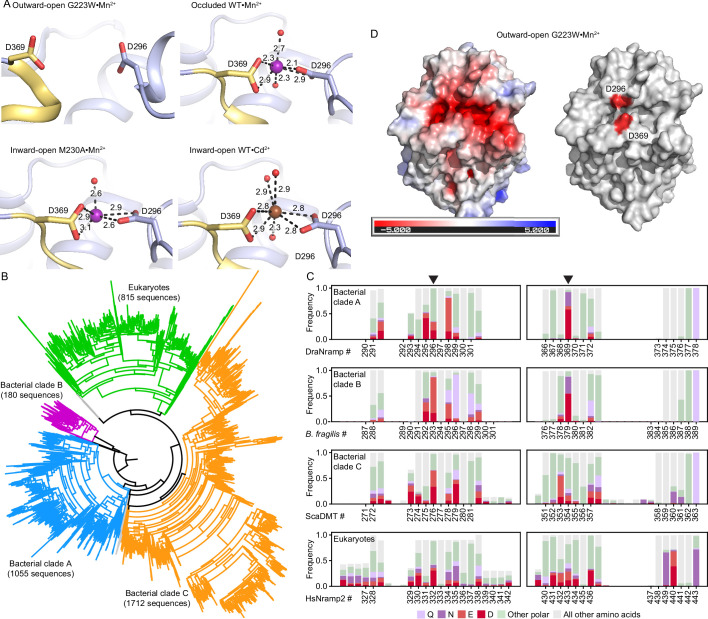
(A) Cartoon representation of WT•Mn2+ in an occluded state. Anomalous signal confirmed the presence of Mn2+ in both the orthosteric metal-binding site and an additional site at the mouth of the external vestibule (Figure 1—figure supplement 1A) which is less conserved across the Nramp family (Figure 1—figure supplement 3). TM1 and TM6 are labeled. (B) Detail of the orthosteric metal-binding site of WT•Mn2+ where D56, N59, M230, and the pseudo-symmetrically related carbonyls of A53 and A227 coordinate the Mn2+ ion (Figure 1—figure supplement 1B). A water molecule completes the six-ligand coordination sphere. Coordinating residues are shown as sticks, and coordinating distances are indicated in Å. (C) ITC measurement of the affinity of WT DraNramp for Mn2+. Top graph shows heat absorbed upon injection of Mn2+ solution to the protein solution. Bottom graph shows the fit of the integrated and corrected heat to a binding isotherm. The data show an endothermic mode of binding and fits best with a two-site sequential binding model. The figure shows one of three measurements and the average Kd values ± SEM (Kd1=190±30 µM, Kd2=1970±520 µM; see Appendix 1). Based on ITC experiments comparing Mn2+ binding to WT or DraNramp constructs with mutations at the external site (Figure 1—figure supplement 2A), we assigned Kd1 to the orthosteric site. In all figures, unless otherwise noted, TMs 1, 5, 6, and 10 are pale yellow, TMs 2, 7, and 11 gray, TMs 3, 4, 8, and 9 light blue, and Mn2+ atoms are magenta spheres.