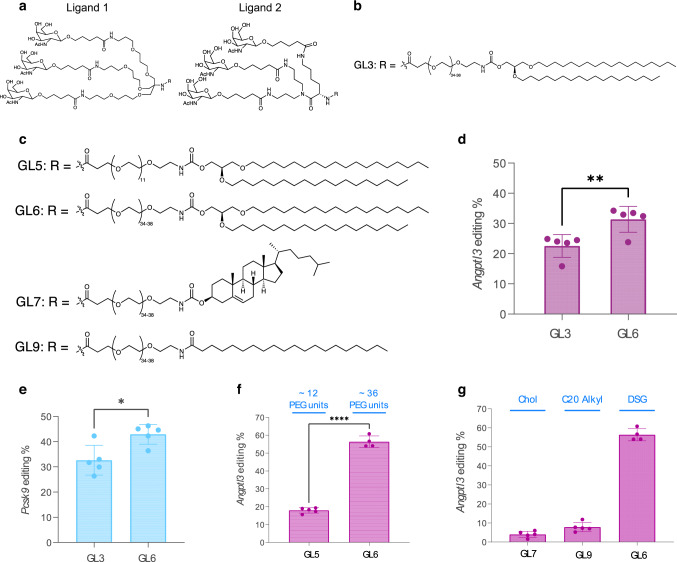
Fig. 1. Structures and initial screen of GalNAc-Lipids.
a Structure of ligand Designs 1 and 2. b Structure of R moiety for GL3, which uses ligand Design 1. c Structures of R moiety for GL5, GL6, GL7, and GL9. These structures utilize ligand Design 2 but differ in their lipid anchors and polyethylene glycol (PEG) spacer lengths. d GalNAc-LNPs constituted with 0.05 mol % GL3 (ligand Design 1 – Table S1, entry 2) or 0.05 mol % GL6 (ligand Design 2 – Table S1, entry 1) were prepared via the in-lipid mixing method and were administered to female 8-10 week old Ldlr –/– mice via injection into the retro-orbital sinus at a dose of 0.1 mg/kg. GL3 and GL6 differ only in the ligand design; the PEG spacer and lipid chain are the same. Ligand 2-based GL6 GalNAc-LNPs achieved higher Angptl3 liver editing when mice were administered GalNAc-LNPs encapsulating mouse-specific Angptl3 guide RNA and ABE8.8 mRNA. Data was analyzed with a two-tailed unpaired T test, p = 0.0086 (df=8, mean difference = 8.9%, 95% confidence interval 2.9–14.7%). e GalNAc-LNPs constituted with 0.5 mol % GL3 (Table S1, entry 4) or GL6 (Table S1, entry 3) were prepared via the post-addition method and were administered to female Ldlr –/– mice via injection into the retro-orbital sinus at a dose of 0.25 mg/kg. Ligand Design 2-based GL6 GalNAc-LNPs achieved higher Pcsk9 liver editing, when mice were administered GalNAc-LNPs encapsulating mouse-specific Pcsk9 guide RNA and ABE8.8 mRNA (N = 5). Data were analyzed with a two-tailed unpaired T test, p = 0.01 (df = 8, mean difference = 10.3%, 95% confidence interval 2.9–17.6%). f GalNAc-LNPs formulated with the longer PEG spacer of GL6 (Table S1, entry 6) achieved higher editing of Angptl3 than the GalNAc-LNPs with the shorter PEG spacer of GL5 (Table S1 entry 5) at 0.3 mg/kg in female Ldlr –/– mice. Data were analyzed with a two-tailed unpaired T test, p < 0.0001 (df = 7, mean difference = 38.5%, 95% confidence interval 34.7–42.2%). g Modulation of the lipid tail hydrophobicity in GL7 and GL9 (Table S1, entries 7 and 8) with cholesteryl (Chol) and arachidoyl (C20) moieties was unable to improve the editing efficiency of GalNAc-LNPs in female Ldlr –/– mice compared to the 1,2-O-dioctadecyl-sn-glyceryl (DSG)-based GL6 (Table S1, entry 6) at 0.3 mg/kg. Data are presented as mean values + /- standard deviation, and individual data points for each animal are displayed (n = 5). * denotes p < 0.05, ** denotes p value <0.01, **** denotes p < 0.0001. Source data are provided as a Source Data file.