Figure 4.
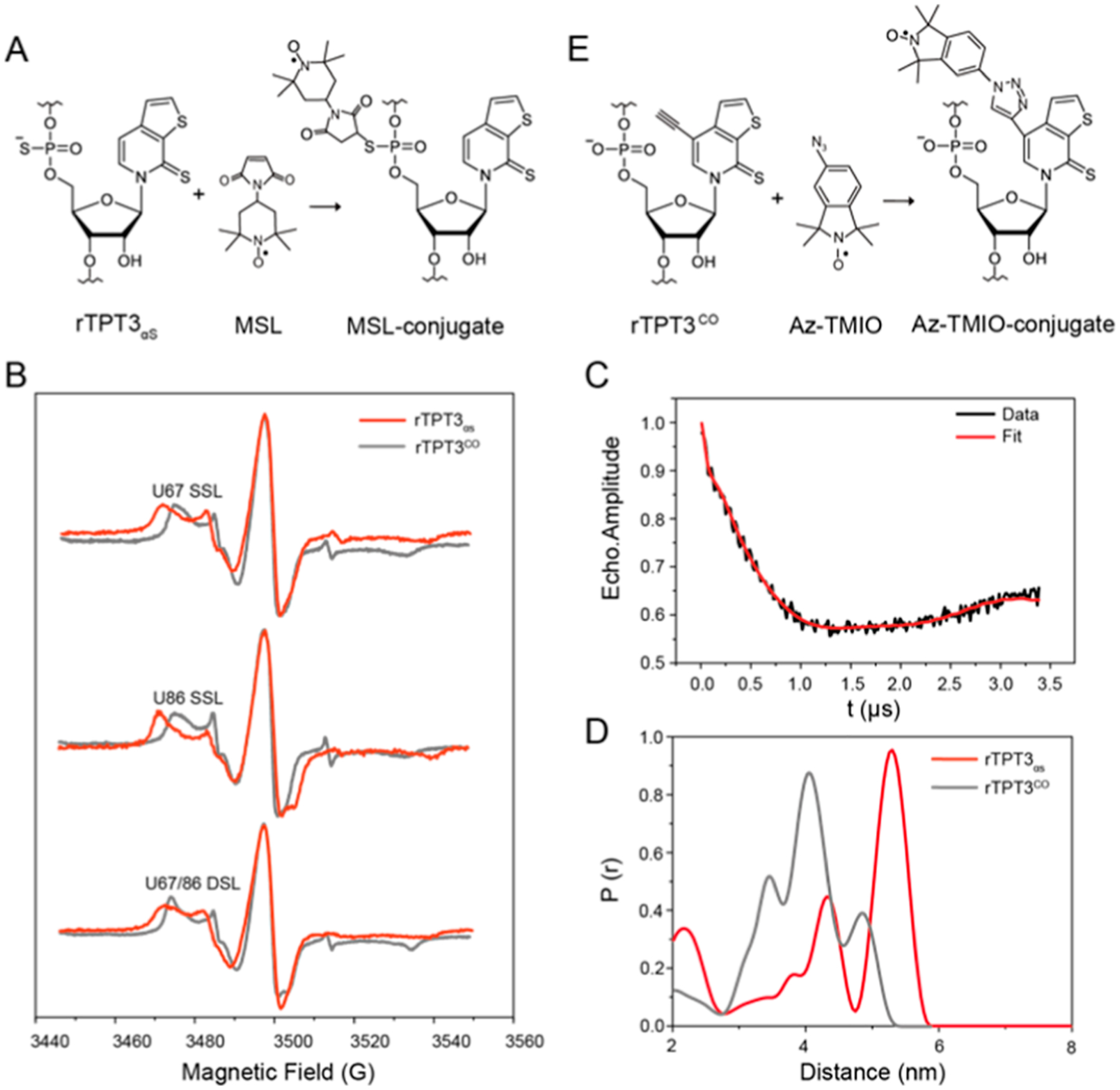
PS-based site-specific spin labeling of RNase P RNA and distance distribution measurement by PELDOR. See also Figure S7 for additional PELDOR data and analysis. (A) Schematic for covalent conjugation of MSL to rTPT3αS-modified RNA via the thiol−maleimide reaction. (B) CW-EPR spectra of the rTPT3αS-based (red) U67 (top) or U86 (middle) SSL and U67/U86 (bottom) DSL RNase P RNAs. CW-EPR spectra of rTPT3CO-based (gray) singly and DSL RNase P RNAs at the same sites were overlaid for comparison. (C) Background-corrected time trace of DSL RNase P. (D) Interspin distance distributions of the rTPT3αS-based (red) and rTPT3CO-based (gray) U67/U86 DSL RNase P RNAs obtained using the Tikhonov regularization approach. (E) Schematic for the click chemistry reaction between RNA containing alkyne-functionalized rTPT3 and azide-modified nitroxide.
