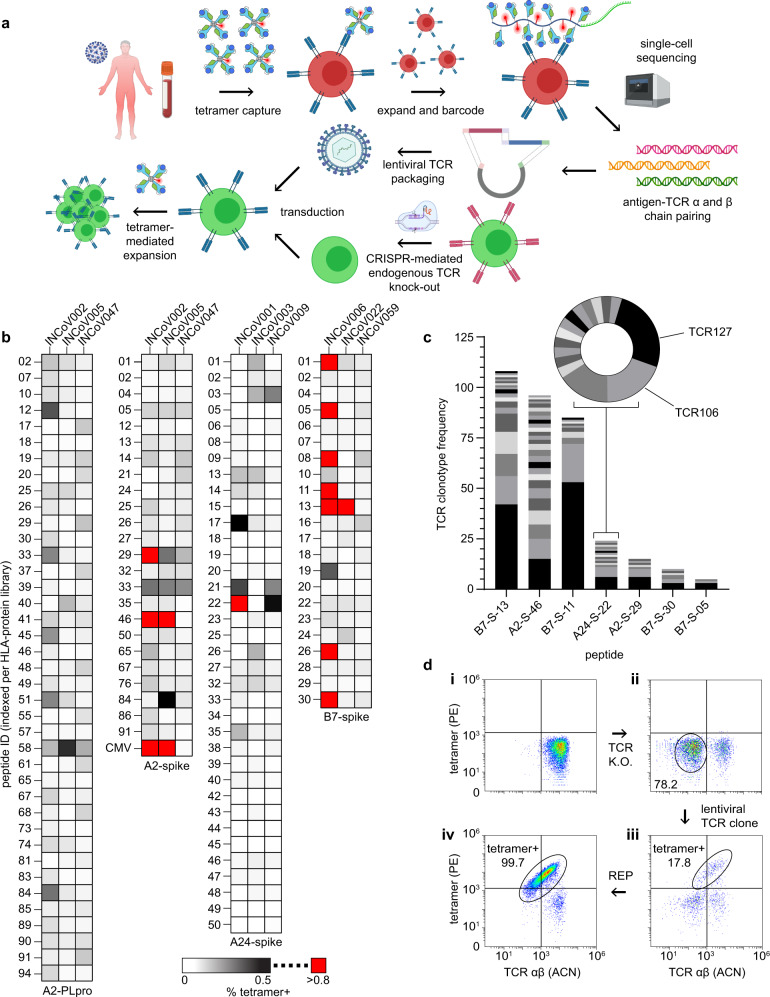
Fig. 4. Isolation and cloning of TCRs specific to SARS-CoV-2 epitopes across three Class I HLA alleles.
a Workflow for SCT-facilitated capture of SARS-CoV-2 antigen-specific CD8+ T cells, single-cell TCR sequencing, and TCR cloning into autologous T cells (created with BioRender.com). b SCT tetramer-positive CD8+ T cells from HLA-matched COVID participants for SCT libraries (n = 1). c Frequency of unique TCR clonotypes against peptides whose SCTs produced high % tetramer binding (red boxes of (b)). d Representative flow cytometry plots of T cell cloning workflow. Autologous T cells (d.i), T cells after CRISPR-mediated TCR knockout (d.ii), tetramer+ T cells after lentiviral TCR cloning (d.iii), tetramer+ T cells after sort and REP (d.iv). Peptide sequences are found in Supplementary Tables 3–6. A2 A*02:01, A24 A*24:02, B7 B*07:02, PLpro papain-like protease, P PLpro, S spike, PE phycoerythrin, ACN allophycocyanin, REP rapid expansion protocol.