Figure 2.
A unique structure in VPS35L regulates VPS29 interaction
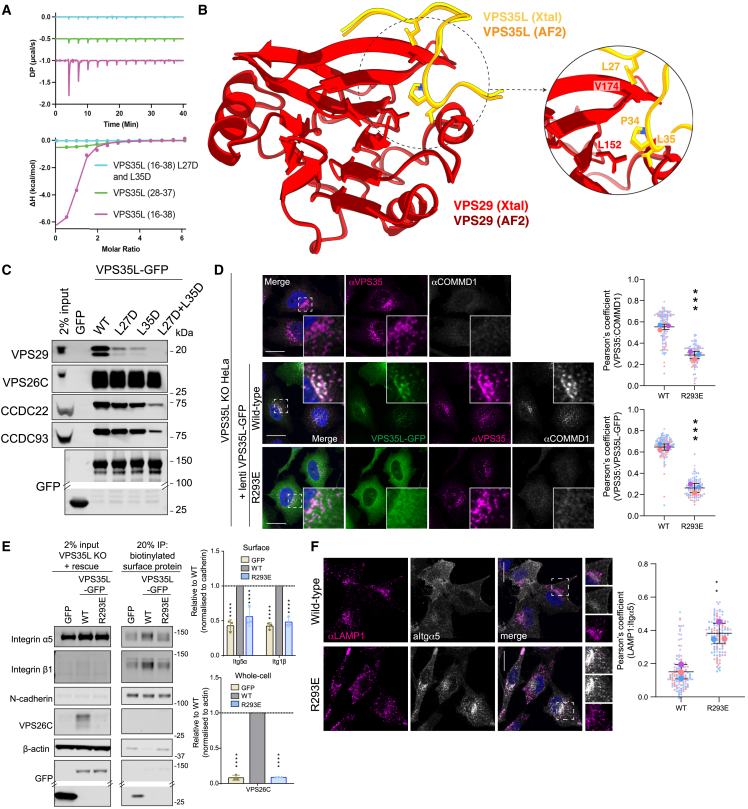
(A) VPS35L peptides were titrated into VPS29 and binding affinity measured by ITC. Top shows the raw data and bottom shows the integrated and normalized data fitted with a 1:1 binding model. VPS35L (16-38) had a slightly higher affinity (1.87μM ± 0.8 μM) than VPS35L (28-37) (6.8μM ± 1 μM), while the L27D/L35D mutant peptide showed no binding. Kd values and standard error of the mean (SEM) are calculated from n = 3.
(B) A 1.35-Å crystal structure of VPS29 bound to VPS35L (16-38) confirms the binding of the core 34PL35 motif to VPS29 and extended interaction of adjacent residues predicted by AlphaFold2.
(C) GFP-nanotrap of GFP-VPS35L mutants in the 26PL27 and 34PL35 sequences. Data S1 shows quantified and raw blots (n = 3).
(D–F) Expression of VPS35L(R293E) in a VPS35L knock-out HeLa cells fails to: (D) rescue the localization of VPS35L or the CCC complex to Retromer-decorated endosomes as observed with wild-type VPS35L; (E) the expression and stability of VPS26C and the steady-state cell surface level of α5β1-integrin; (F) the trafficking of α5-integrin away from LAMP1-positive late endosomes/lysosomes. Data S1 shows quantified band intensities and raw blots. (D and F) Pearson’s coefficients were quantified from >30 cells per 3 independent experiments. Pearson’s coefficients for individual cells and means are presented by smaller and larger circles, respectively, colored according to the independent experiment. The means (n = 3) were compared using a two-tailed unpaired t test. Error bars represent the mean, S.D. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figure S1.