Figure 5.
DENND10 associates with the central coiled-coil domains of CCDC22 and CCDC93
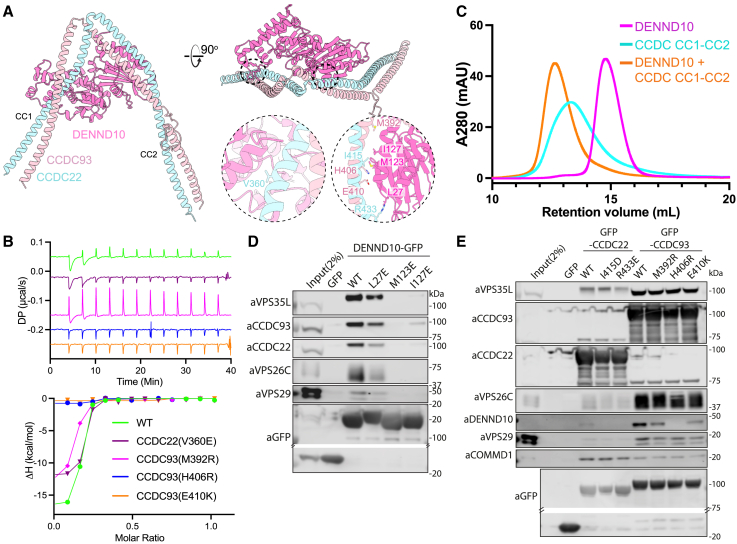
(A) Structure of DENND10 complex with the dimeric CC1 and CC2 coiled-coil regions of CCDC22 and CCDC93 predicted by AlphaFold2. Model quality and predicted alignment errors are shown in Figure S6.
(B) DENND10 was titrated into purified wild-type and mutant CC1-CC2 complexes (CCDC22(325–485) + CCDC93(310–488)) and binding was measured by ITC. Top shows the raw data and bottom shows the integrated and normalized data fitted with a 1:1 binding model. The binding affinities were as follows: WT, 34 nM ± 0.5 nM; CCDC22 (V360E), 47.9 nM ± 3.5 nM; and CCDC93 (M392R), 89.1 nM ± 10 nM. No binding was detected for CCDC93H406R or E410K.
(C) Analytical size exclusion chromatography of DENND10 (magenta), CC1-CC2 complex (cyan), and DENND10 mixed with CCDC22-CCDC93 forming a stable complex (orange).
(D and E) GFP-nanotrap of GFP-DENND10 (D) or CCDC22 and CCDC93 mutants. Data S1 shows quantified band intensities and raw blots (n = 3).
See also Figure S6.