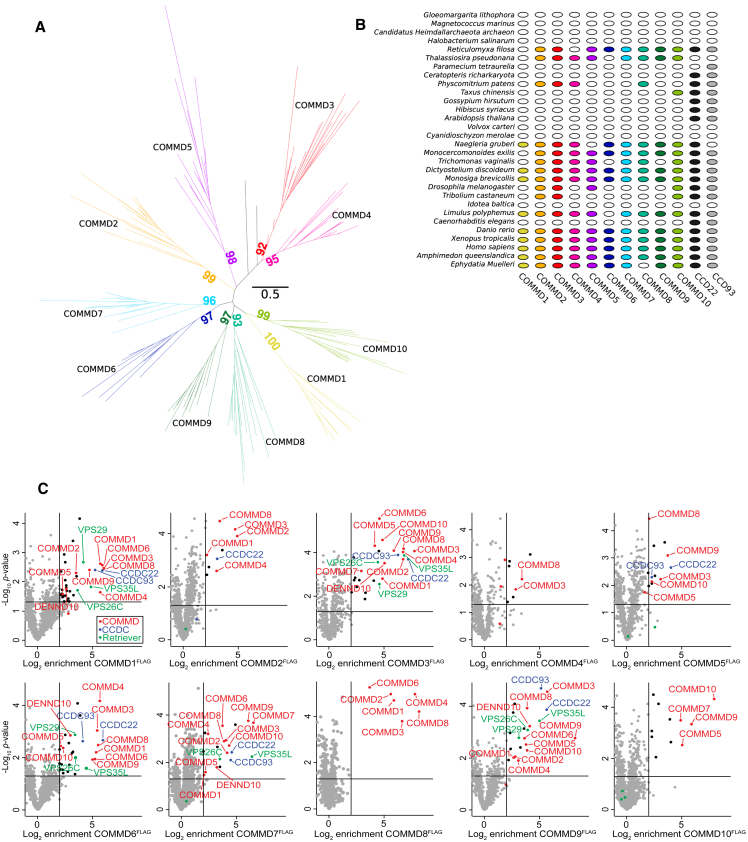
Figure S5.
Evolutionary origins of the CCC complex, related to Figure 4
(A) Maximum-likelihood phylogeny of COMMD1-10 proteins from 23 representative eukaryotic taxa inferred under the best-fitting Q.yeast+R5 substitution model. Each COMMD forms a strongly supported (>90% bootstrap) clan in the unrooted phylogeny, and each clan contains representatives from all major lineages (supergroups) of eukaryotes; this implies that all ten COMMD subunits were already present in the last eukaryotic common ancestor (that is, LECA appears 10 times in the tree). Based on the absence of COMMD homologues in Bacteria and Archaea, this protein family likely originated on the eukaryotic stem and proliferated via a series of gene duplications prior to the radiation of the modern eukaryotic groups.
(B) Presence-absence patterns of COMMD and CCDC22/CCDC93 in a set of representative modern eukaryotes. The presence-absence pattern of COMMD genes in modern eukaryotes, taken together with the phylogeny in (A), indicates that these genes have been lost independently in different eukaryotic lineages.
(C) Digitonin-solubilized COMMD knockout eHap cell lines expressing the indicated COMMDFLAG construct were affinity enriched using Flag agarose beads followed by label free quantitative proteomics. The threshold of significant enrichment was determined to be 2-fold (log2 fold change = 1) based on the distribution of unenriched proteins. Black and colored dots indicate significantly enriched proteins. Red, COMMD subunits; Blue, CCDC subunits; Green, Retriever subunits.