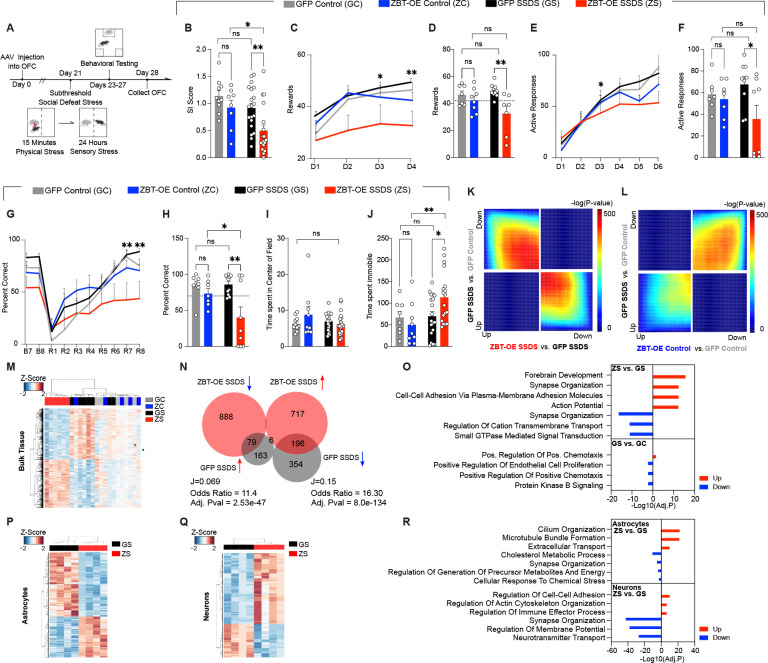
Fig. 4. ZBTB7A in mouse OFC astrocytes is sufficient to induce chronic stress-mediated alterations in chromatin accessibility, gene expression, and behavior.
(A) Schematic of experimental timeline with subthreshold SSDS mild stress paradigm performed after rAAV6 injection into OFC, followed by behavioral tests and tissue collection for RNA-seq. (B) Social interaction. 2-way ANOVA main effect of stress [F1,52 = 8.144], **p = 0.0062, main effect of virus [F1,52 = 7.730], **p = 0.0075. Sidak’s MC test, GFP control vs. GFP SSDS ns, p = 0.2788. GFP SSDS vs. ZBT-OE SSDS **p = .0041. ZBT-OE control vs. ZBT-OE SSDS *p = .0286. GFP control vs. ZBT-OE control n.s. p = .4480. (C) Pavlovian cue-reward association task. “D” indicates Day of test. 3-way ANOVA, main effect of Virus x Stress [F1,29 = 5.291] *p = 0.0288. (D) Individual values for day 2 of task shown in (C). 2-way ANOVA, main effect of virus [F1,28 = 9.759], p = **0.0041. Sidak’s MC test, GFP control vs. GFP SSDS ns, p=0.651. GFP SSDS vs. ZBT-OE SSDS **p = .0021. ZBT-OE control vs. ZBT-OE SSDS ns, p = 0.146. (E) Operant reward task, FR1. “D” indicates Day of test. 3-way ANOVA, main effect of Test Day x Virus [F5,149 = 2.823] *p = 0.0182. (F) Individual values for day 3 of task shown in (E). 2-way ANOVA, main effect of virus [F1,27 = 4.408], *p = 0.0453. Sidak’s MC test, GFP control vs. GFP SSDS ns, p=0.709. GFP SSDS vs. ZBT-OE SSDS *p = .0218. ZBT-OE control vs. ZBT-OE SSDS ns, p = 0.282. (G) Percent correct trials in reversal learning paradigm. “B” indicates Baseline day, “R” indicates Reversal phase day. 3-way ANOVA, main effect of Test day x Virus [F9,261 = 4.529] p < 0.0001. (H) Individual values for day 7 as shown in (G). 2-way ANOVA, main effect of virus [F1,30 = 9.017], **p = 0.0054. Sidak’s MC test, GFP control vs. GFP SSDS ns, p=0.9797. GFP SSDS vs. ZBT-OE SSDS **p = .0013. ZBT-OE control vs. ZBT-OE SSDS *p = 0.0389. GFP control vs. ZBT-OE control n.s., p = 0.7280. (I) Time spent (s) in the center of the field during open field test. 2-way ANOVA ns, (J) Forced Swim tests. 2-way ANOVA main effect of interaction [F1,50 = 4.129], *p = 0.0475, main effect of stress [F1,50 = 4.993], *p = 0.0475. Sidak’s MC test, GFP control vs. GFP SSDS ns, p=0.9876. GFP SSDS vs. ZBT-OE SSDS **p = 0.0070. (K-L) RRHO comparing gene expression between indicated comparisons, in the context of mild stress. (M) Clustering at 1,929 DE genes between ZBT-OE SSDS and GFP SSDS. (N) Scaled Venn-diagram and odds ratio test of the overlap between differentially expressed (DE) genes in bulk OFC tissues comparing ZBT-OE stress vs. GFP SSDS, with GFP SSDS vs. GFP control. “J” indicates the Jaccard index. Note for GFP SSDS vs. GFP control, DEGs were defined at pval < 0.05 (O) GO analysis for gene DEGs in ZBT-OE SSDS vs. GFPSSDS and GFP SSDS vs. GFP control, separated by up/down regulation. All data graphed as means ± SEM. (P) Clustering at 715 DE genes between ZBT-OE SSDS and GFP SSDS astrocytes (n = 4/group). (Q) Clustering at 1,191 DE genes between ZBT-OE SSDS and GFP SSDS neurons (n = 4/group). (R) GO analysis for DE genes (FDR < 0.1) between ZBT-OE SSDS and GFP SSDS groups in MACs-isolated astrocytes and neurons, separated by up/down regulation.