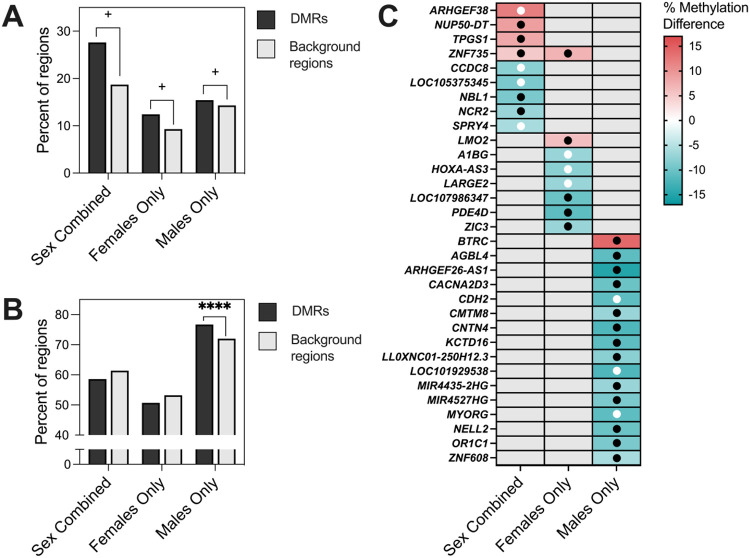
Figure 6. Comparison of DS-CHD DMRs with DS vs TD samples.
A) Percent of DS-CHD DMRs and background regions that were significantly differentially methylated in DS vs TD samples. Z-test for two population proportions, Sex Combined (z =1.7343, two-tailed p =1.08364) Females Only (z =1.93, two-tailed p =0.0536), Males Only (z =1.8808, two-tailed p =0.0601). + = p <0.1. B) Percent of DS-CHD DMRs that were methylated in same direction in DS vs TD as in DS-CHD vs DS non-CHD. Z-test for two populations proportions, Sex Combined (z =−0.4274, two-tailed p =0.6672), Females Only (z =−0.8936, two-tailed p =0.37346), Males Only (z = 6.5357, two-tailed p <0.00001). **** = p <0.00001. C) Heatmap showing DS-CHD DMRs that were significant (q < 0.05) in DS vs TD samples mapped to genes. Red indicates hypermethylation in CHD compared to non-CHD while blue represents hypomethylation, with stronger shades representing a greater percent methylation difference and gray meaning that that DMR was not significant for that comparison. Black dots indicate that methylation is in the same direction for DS vs TD as DS-CHD vs DS non-CHD while white dots indicate methylation is in the opposite direction.