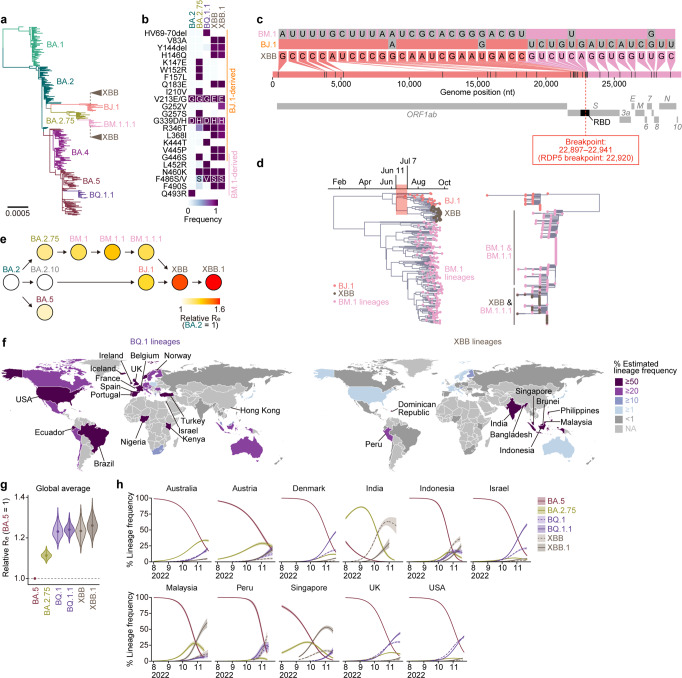
Fig. 1. Phylogenetic and epidemic analyses of the XBB lineage.
a Muximum likelihood tree of representative sequences from PANGO lineages of interest: BA.1, BA.2, BA.4, BA.5, BA.2.75, BJ.1 and BM.1.1.1, rooted on a B.1.1 outgroup (not shown). The recombinant parents of XBB are annotated on the tree as cartoon clades. b Amino acid differences in the S proteins of Omicron lineages. c Nucleotide differences between the consensus sequences of the BJ.1, BM.1 (including BM.1.1/BM.1.1.1) lineages and the XBB (including XBB.1) lineage, visualized with snipit (https://github.com/aineniamh/snipit). d Maximum clade credibility time-calibrated phylogeny of the 5ʹ non-recombinant segment (1–22,920) of the XBB variant (left) and non-calibrated maximum likelihood phylogeny of the 3ʹ non-recombinant segment (22,920–29,903) (right). The right hand-side tree is rooted on a BA.2 outgroup (not shown). e Relative effective reproduction number (Re) values for viral lineages in India, assuming a fixed generation time of 2.1 days. The Re of BA.2 is set at 1. Dot color indicates the posterior mean of the Re, and an arrow indicate phylogenetic relationship. See also Supplementary Fig. 1b. f Difference in the circulated regions between BQ.1 and XBB lineages. Estimated lineage frequency as of November 15th, 2022 in each country is shown. Countries with ≥50% and ≥20% frequencies are annotated for the BQ.1 and XBB lineages, respectively. g Relative Re values for viral lineages, assuming a fixed generation time of 2.1 days. The Re value of BA.5 is set at 1. The posterior (violin), posterior mean (dot), and 95% Bayesian confidential interval (CI; line) are shown. The global average values estimated by a hierarchical Bayesian model27 are shown. See also Supplementary Fig. 1c. h Estimated lineage dynamics in each country where BQ.1 and XBB lineages cocirculated. Posterior mean, line; 95% CI, ribbon. Source data are provided with this paper.