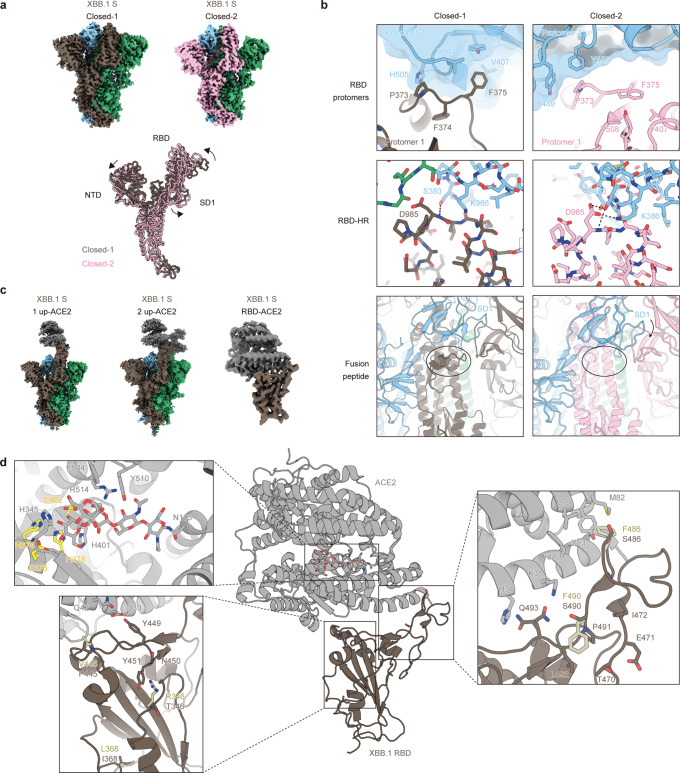
Fig. 4. Overall cryo-EM structure of XBB.1 S and ACE2.
a (Top) Cryo-EM maps of XBB.1 spike (S) protein trimer closed-1 state (left) and closed-2 state (right). Each protomer is colored brown, blue, green (closed-1) or pink, blue, green (closed-2). (Bottom) Superimposed structures of XBB.1 S protomers between closed-1 state (brown) and closed-2 state (pink). b Close-up and corresponding views of the closed-1 and closed-2 structures (same colors as m). (Top) A loop containing F375 at the protomer interface in the receptor binding domain (RBD) region. Each adjacent protomer is shown with the surface model (transparent, blue). (Middle) Interfaces between RBD and heptad repeat-1 (HR-1). Dashed lines represent hydrogen bonds. (Bottom) Structural difference around fusion peptides (shown in cartoon) surrounded by a circle. c Cryo-EM maps of XBB.1 S protein (same colors as m) bound to angiotensin-converting enzyme 2 (ACE2) (gray) in one-up state (left), two-up state (middle), or RBD-ACE2 interface (right). d Structure of RBD-ACE2 complex (same colors as o). In close-up views, corresponding five residues in the BA.2.75 RBD-ACE2 complex structure (PDB: 8ASY)96 different from that of XBB.1 (brown stick) are shown in pastel yellow sticks. Residues interacting with these five amino acid residues in the XBB.1 or the BA.2.75 RBD, as well as residues recognizing the N103-linked glycan of ACE2, are represented by stick models. Residues of the HEXXH motif in the active site of ACE2 are highlighted in yellow.