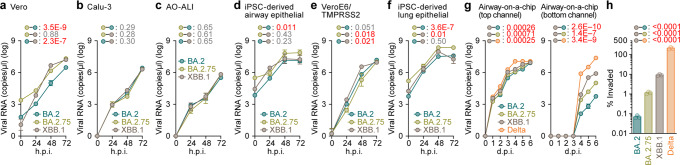
Fig. 5. Growth kinetics of XBB.1.
Clinical isolates of BA.2, BA.2.75, XBB.1 and Delta (only in g, h) were inoculated into Vero cells (a), Calu-3 cells (b), the human airway organoid-derived air-liquid interface (AO-ALI) system (c), human induced pluripotent stem cell (iPSC)-derived airway epithelial cells (d), VeroE6/TMPRSS2 cells (e), iPSC-derived lung epithelial cells (f) and an airway-on-a-chip system (g). The copy numbers of viral RNA in the culture supernatant (a, b, e), the apical sides of cultures (c, d, f), and the top (g, left) and bottom (g, right) channels of an airway-on-a-chip were routinely quantified by RT–qPCR. In (h), the percentage of viral RNA load in the bottom channel per top channel at 6 days post-infection (d.p.i.) (i.e., % invaded virus from the top channel to the bottom channel) is shown. Assays were performed in triplicate (g, h) or quadruplicate (a–f). The presented data are expressed as the average ± standard error of mean (SEM). In (h), each dot indicates the result of an individual replicate. In (d–k), statistically significant differences across timepoints were determined by multiple regression. In (h), statistically significant differences versus XBB.1 were determined by two-sided Student’s t tests. The familywise error rates (FWERs) calculated using the Holm method (a–g) or P values (h) are indicated in the figures. Source data are provided with this paper.