Fig. 6. Promiscuous expression of innate lymphoid genes in ΔDNSe DN3 cells.
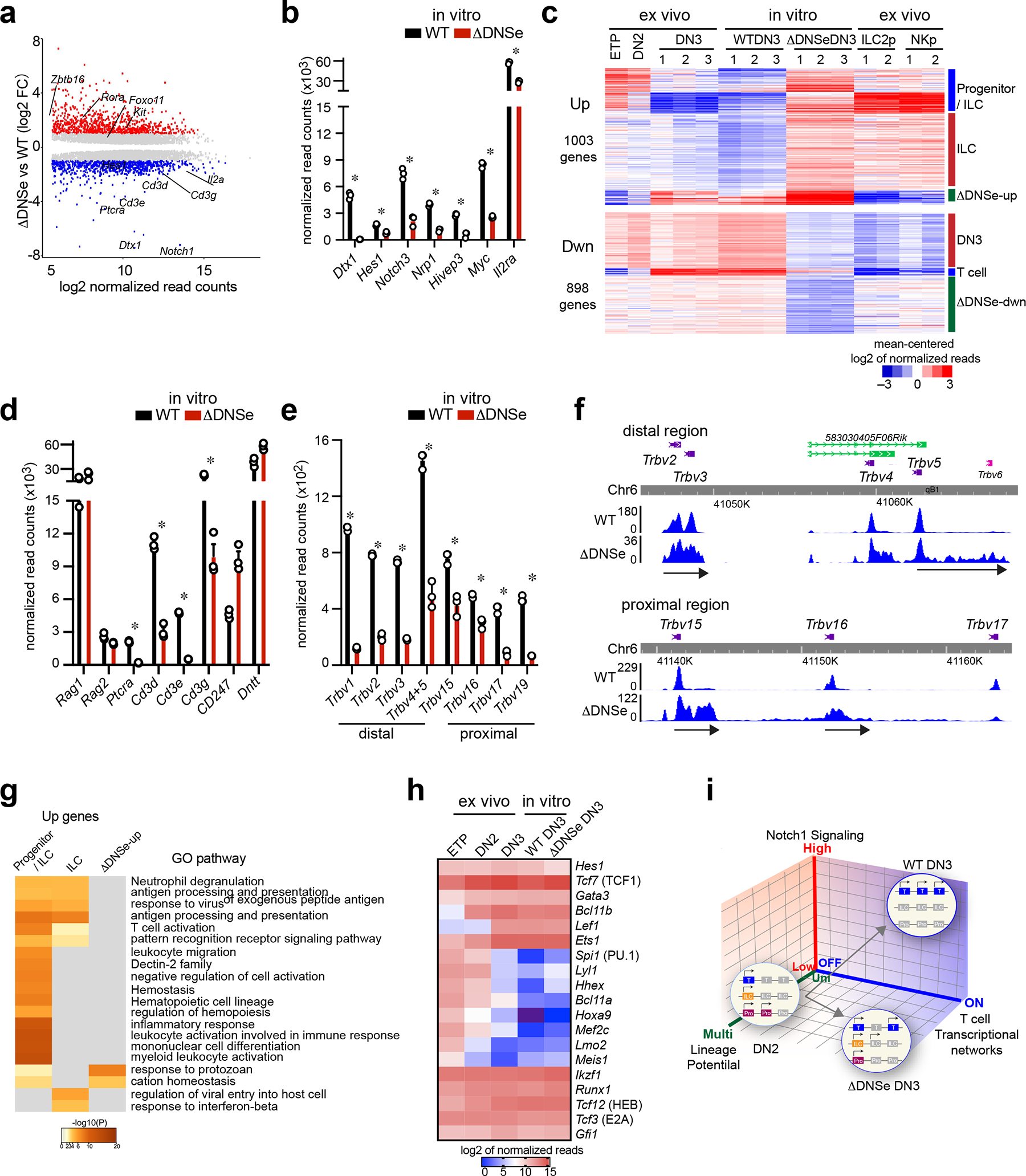
(a) Differentially expressed genes (mean normalized read counts>50, adj. p <0.05) are shown for cultured ΔDNSe vs WT DN3. Up- (log2FC>1, adj. p <0.05, 1003 genes) and Down- (log2FC <−1, adj. p <0.05, 898 genes) regulated genes are highlighted in red and blue, respectively. (b) Expression of Notch1 downstream target genes in cultured DN3 cells is shown (mean+/−sem). * ΔDNSe vs WT DN3: log2FC <−1 or >1, adj. p <0.05 (c) Heatmap of K-means clustering for Up- and Down-regulated genes in cultured ΔDNSe vs WT DN3 is provided. Deregulated genes were co-clustered with expression data from ex vivo WT T cell progenitors (ETP, DN2 DN3 cells) and WT innate lymphoid progenitors (ILC2p and NKp). (d) Expression of Rag1, Rag2, Dntt, Ptcra, and Cd3 genes in cultured DN3 cells is shown (mean+/−sem). * ΔDNSe DN3 vs WT DN3: log2FC <−1 or >1, adj. p <0.05 (e and f) Expression of Trbv genes is shown by bar graph (mean+/−sem, * adj. p <0.01) (e) and RNAseq read distribution is visualized by genome browser (f). (g) Heatmap based on significance of functional pathway enrichment (−log10(pval)) is shown for up-regulated genes classified into three signatures as in (c). (h) Heatmap of gene expression for key transcription factors in T cell development is provided. Analysis of combined datasets from ex vivo WT DN3 and cultured DN3 from WT and ΔDNSe is shown from three independent samples. (i) A model on how strength of Notch1 signaling controls induction of T cell transcriptional networks and lineage restriction is provided. Genes associated with the T cell lineage, early progenitors, or innate lymphocytes are depicted by blue, purple, and orange boxes, respectively. Silenced genes are shown in gray.
Data for cultured DN3 cells were generated from three independent experimental groups for each genotype. Data for ex vivo WT ETP and DN2 were generated from one experimental group and for ex vivo WT DN3 from three independent experimental groups as in Fig. 1. Data for the two replicates of ILC2p and NKp were obtained from published datasets (GEO: GSE77695).
