Extended Data Fig. 1. Generation of mice with deletion of the DNS region and effects of the DNS and Notch1 deletions on gene expression in DN3 thymocytes.
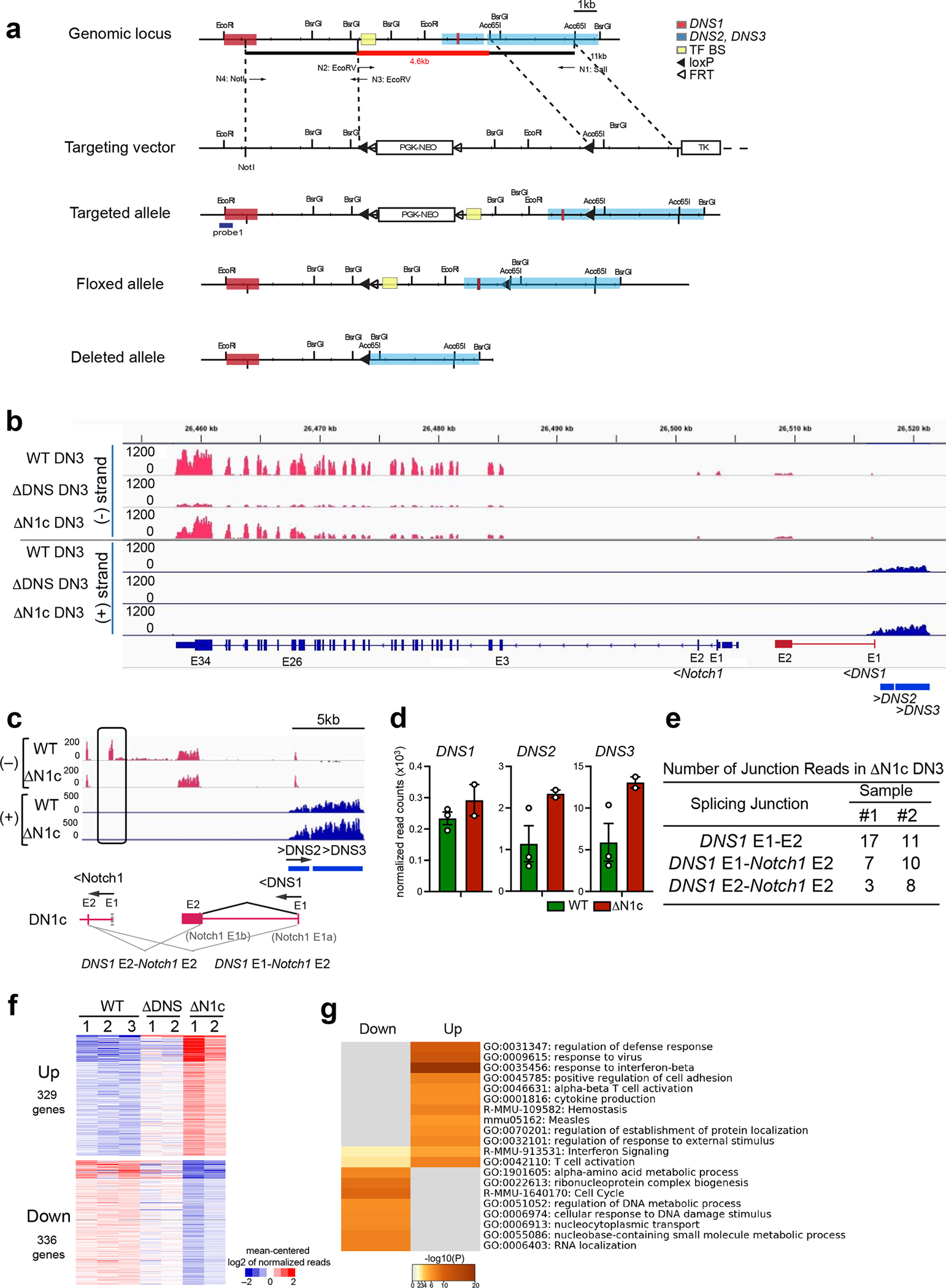
(a) Strategy to generate a conditional deletion of the DNS region. A 4.6 kb region containing the DNS promoter and a region enriched for transcription factor binding sites (TF BS) were flanked by loxP sites (black arrowhead). The Neo marker was flanked by Frt sites (white arrowhead). Flp recombinase excises the Neo gene from the targeting construct and generates the DNS floxed allele. CD2-Cre or Mx-Cre recombinase were used to induce deletion of the DNS region in T cell progenitors or in all hematopoietic cells and progenitors, respectively. DNS1 exons (red square) and DNS2 and DNS3 exons (blue square) and TF BS (yellow square) are shown. (b, c) Genome browser tracks of normalized RNAseq reads at Notch1 ((−) strand), DNS1 ((−) strand), and DNS2 and DNS3 ((+) strand) genes in WT, ΔDNSCD2cre, and ΔN1cCD2cre DN3 thymocytes are shown. Complete loss of Notch1 exon1 (E1) was detected in ΔN1cCD2cre DN3 (b, c). Splicing events in ΔN1cCD2cre are drawn by angled lines (c). (d) Expression of DNS1-3 RNAs in DN3 is shown (mean +/− SEM). Data shown for WT DN3 is also used in Fig. 1g. (e) Number of RNA-seq reads that span splicing junctions are shown for two independent ΔN1cCD2cre DN3 data sets. (f) K-means clustering of genes significantly up-or down- regulated in ΔN1cCD2cre vs WT (log2FC>0 or <0, adj.p <0.05) in WT, ΔDNSCD2cre, and ΔN1cCD2cre DN3 cells is shown. A similar trend of up- and down-regulated genes was seen with ΔDNSCD2cre and ΔN1cCD2cre DN3 cells relative to WT DN3 cells although gene expression changes observed in ΔDNSCD2cre were smaller than those seen in ΔN1cCD2cre DN3. (g) GO Pathway analyses of up- or down-regulated genes in ΔN1cCD2cre vs WT.
Data shown were generated from two (ΔDNSCD2cre DN3 and ΔN1cCD2cre DN3) and three (WT DN3) independent experimental groups with pooled samples from at least two biological replicates.
