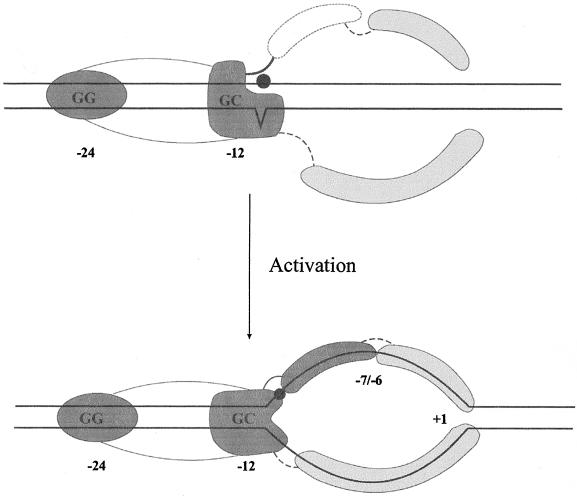
FIG. 3.
Cartoon model showing activities proposed for activation via DNA melting. Dark shading indicates activities primarily in ς54. These activities include duplex (double-stranded DNA) binding at nucleotides −24 and −12 and fork junction binding. The −7 to −11 single-strand binding activity is seen only after activation. Light shading indicates single-strand binding activities contributed by RNA polymerase holoenzyme. The solid circle represents the −11 connector nucleotide that inhibits the spread of unactivated melting when unpaired. (Top) Prior to activation, the holoenzyme is stabilized on the DNA using primary interactions at nucleotide −24. The unactivated complex includes a form shown here in which a molecular structure near −12 contributes to binding and prevents the spread of early DNA melting to downstream positions. The N terminus and the fork junction connector position −11 (solid circle) are key determinants in locking the system in a closed state. Deregulated bypass mutations destroy this ς54-DNA structure at −11 and allow partial engagement of the downstream single-strand binding activities; these are shown downstream of −12 and above and below the DNA in an unengaged state. (Bottom) Upon activation, ATP triggers conformational changes in ς54 at the fork junction that involve the ς54 N terminus. The upstream nontemplate strand binding activity is exposed, and interactions through the connector can now be established. The single-strand DNA-binding activities stabilize the spread of melting.