Figure 1. Overview of More Than 10 Years of Molecular Combing–Based Diagnosis of FSHD.
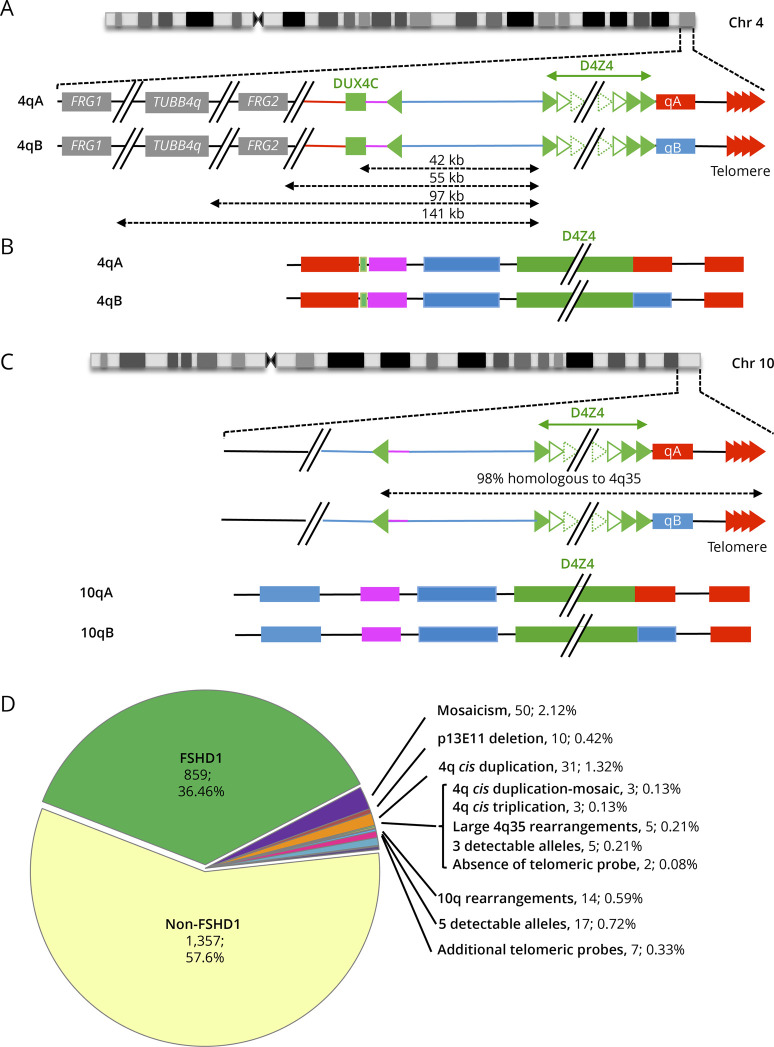
(A) The most distal 4q region is represented (4q35 locus) together with the proximal FRG1, TUBB4Q, and FRG2 genes. As described in reference 16, the D4Z4 array is depicted by green triangles. Sequences starting with an inverted D4Z4 repeat (green arrow) are specific to the 4q35 locus (red lines). Regions located between the D4Z4 array and the inverted D4Z4 repeat are also present on the 10q26 locus (blue bar).The 4qA (red rectangle) is characterized by the presence of the pLAM sequence distal to the last D4Z4 repeat and followed by the telomere (red arrows). The 4qB allele (blue rectangle) differs from the 4qA.16 (B) The V3 pink barcode used to distinguish the different alleles (4qA/B; 10qA/B) is based on a specific combination of 4 different colors (blue, pink, red, and green) used to label the 4q35 locus and 10q26 loci up to the telomeric sequence.21 The proximal 4q-specific region is detected by a combination of red and pink probes. The proximal 10q-specific region is identified by a blue probe. For chromosome 4, the four-color barcode comprises 1 probe detected in blue and one in pink, which hybridize the proximal region, one 6 kb red probe, which hybridizes the (TTAGGG)n telomeric ends (red). The qB-specific probe, adjacent to D4Z4, is detected in blue. (C) Schematic representation of chromosome 10q and illustration of the V3 pink barcode used to distinguish the 2 10q alleles (qA/B) based on a combination of 4 different colors and different DNA probes encompassing the distal regions up to the telomeric sequence. The barcodes for the 4q-10q homologous regions are identical. The proximal 10q-specific region is identified by a blue probe leading to a combination of blue-pink-blue probe for the 10q26 locus and red-pink-blue for the 4q35 region. The distal A-type region is identified by a red probe, the B-type allele by a blue probe.16 (D) Of the 2,363 patients analyzed since 2011 by MC, 91.3% showed a normal profile with 4 distinct alleles and absence (1,357 cases, 57.6%) or presence (859 cases, 36.46%) of D4Z4 array contraction on a 4qA allele. The authors identified 6.26% of cases with an atypical profile with 2.12% (n = 50) of samples with a mosaic D4Z4 array contraction, 0.42% (n = 10) with a deletion of the p13E-11 probe, 1.32% (n = 31) of patients with a 4qA cis-duplication,16 0.13% (n = 3) with a cis-duplication in mosaic, 0.13% (n = 3) with a cis-triplication, and 0.21% (n = 5) with a complex 4q35 rearrangement. In 0.21% (n = 5) of cases, only 3 alleles were detectable while 5 alleles were observed in 0.72% (n = 17) of cases. The authors noticed the presence of 10q26 rearrangements in 0.59% (n = 14) of cases, the absence of telomeric probes in 2 cases (0.08%), and 0.33% (n = 7) with multiple telomeric probes. FSHD = facioscapulohumeral dystrophy; MC = molecular combing.