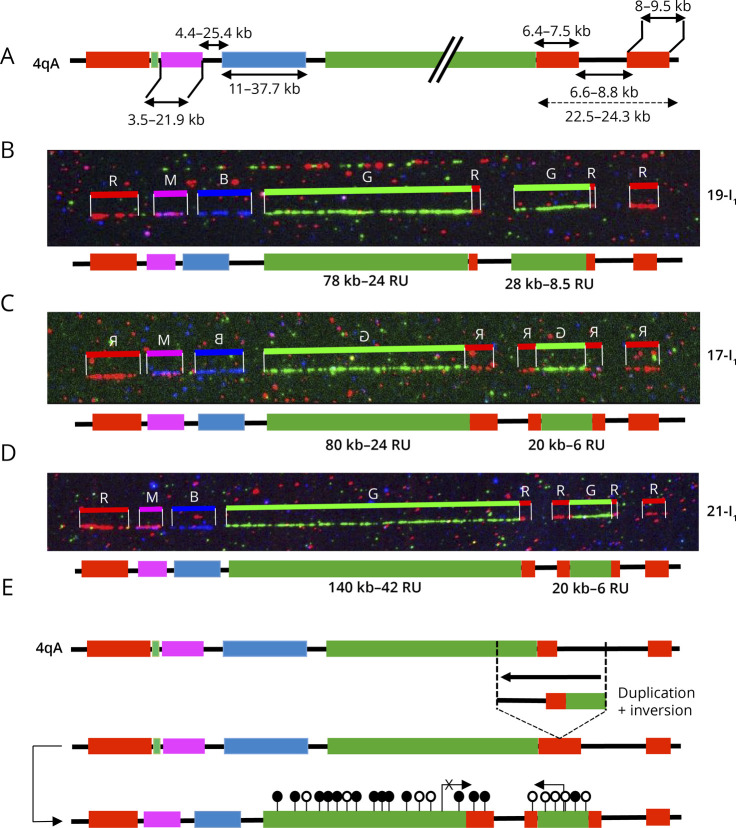
Figure 2. Presence of a Red Probe and Gap Upstream of the Duplicated D4Z4 Array Suggests Genomic Inversion.
(A) Schematic representation of the 4q35 barcode. The size of the different regions was estimated by the analysis of >1000 DNA samples previously processed by MC.7 For the 4qA allele, a probe labeled in red hybridizes the qA-specific β-satellite region of variable lengths (6.4–7.5 kb). The size of the gap between the A-type and telomeric probe is estimated around 6.6–8.8 kb.7 (B–D) Images and schematic representations of new cis-duplication identified using the Fibervision tool. For each D4Z4 array, the size in kb is indicated together with the number of D4Z4 units. (B) Absence of the red probe corresponding to the A-type region upstream of the duplicated D4Z4 array (case 19-I1). (C and D) Presence of the red probe (A-type region) upstream of the duplicated D4Z4 array (C, case 17-I1; D, case 21-I1). (E) Schematic representation of the possible mechanism leading to the 4q35 cis-duplication. The authors hypothesize a duplication and inversion of the region containing a variable number of D4Z4 repeats (<10 RUs), the A-type region, and the gap between this A-type region and the telomere followed by insertion or the duplicated region within the A-type region. Depending on the context (presence or absence of SMCHD1 variant), the long D4Z4 array might remain methylated (black dots) while the short allele might be hypomethylated (white dots). In this configuration, the DUX4 coding region is in the opposite orientation. It remains to be determined whether the recombined allele remains permissible for transcription. MC = molecular combing.