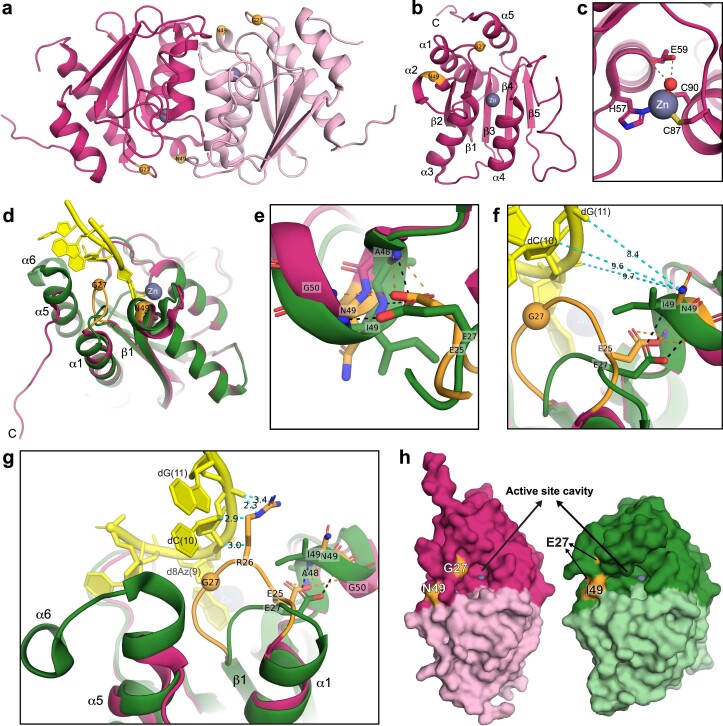
Extended Data Fig. 4. Crystal structure of TADAC-1.19 without ssDNA and structural comparisons with TadA*8.20.
a, Overall structure of the TADAC-1.19 functional homodimer (dark and light pink). E27G and I49N substitutions relative to TadA*8.20 are shown in orange spheres. Zinc ion is shown as a gray sphere. b, Overall structure of TADAC-1.19 monomer. C represents C-terminus. c, TADAC-1.19 active site with water (red sphere) bound to the zinc ion. H57, C87, and C90 coordinate with the zinc ion. The water molecule H-bonds (dashed lines) to the catalytic residue E59. d–h, Structural comparisons between TADAC-1.19 and TadA*8.20 structures. (d) Superposition between substrate-free TADAC-1.19 (pink) and ssDNA-bound TadA*8.20 (green, yellow) monomers, showing high structural similarity (RMSD of ~0.9-Å for all of the Cα atoms). The main structural differences are in α1-helix, the loop between α1 and β1, and C-terminal α5- and α6- helices. (e) TadA*8.20 has E27 side chain H-bonding (black dashed lines) to the main chains of A48, I49, and G50, and E27G substitution removes these interactions. To compensate for these critical contacts, TADAC1.19 places E25 at a similar position to the formerly occupied by E27 in TadA*8.20 to make the same H-bonds (orange dashed lines) with A48, I49, and G50. E25 displacement shortens α1-helix, and the loop between α1 and β1 (orange), containing E27G substitution, is extended and adapts into a different conformation compared to TadA*8.20 (d, f, and g). This results in partial unfolding of α5-helix to prevent steric clash with this loop conformation and complete unfolding of α6-helix (d and g). These structural changes alter the shape of the TADAC-T1.19 active site cavity (h), affecting substrate binding in the active site (Fig. 2b). R26 present in TADAC-T1.19-loop (orange) would make close contacts (cyan dashed lines) with dC(10), adjacent to the target base d8Az(9), and dG(11) of TadA*8.20-ssDNA (g). I49N substitution positions the N49 side chain far from the ssDNA backbone (~9-Å from dG(11); cyan dashed lines) (f), suggesting that a residue with a longer positively charged side chain like lysine would create additional contacts with the ssDNA, as observed in the TADAC-T1.14 structure (Extended Data Fig. 3e).