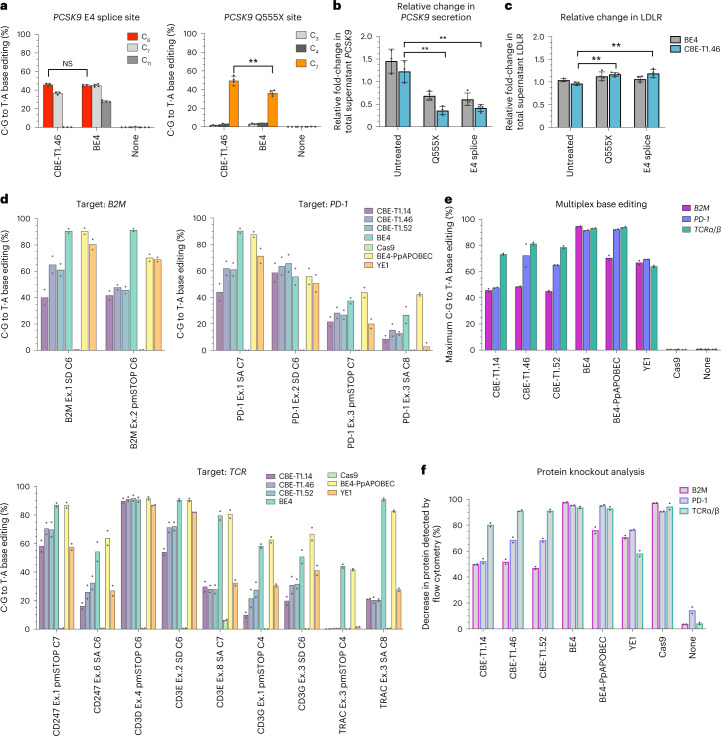
Fig. 6. Evaluation of CABE-Ts and CBE-Ts in therapeutically relevant cell contexts.
a, Gene-editing outcomes of primary human hepatocytes transfected with mRNAs encoding CBE-T1.46 and BE4 at three sites within the PCSK9 gene (left, P = 0.2882 (NS); right, P = 0.0017 (double asterisk)). Positional edit within the protospacer indicated. b, Evaluation of relative change in PCSK9 secretion between day 9 (collection time point) and day 0 (transfection time point) through ELISA. The targeted PCSK9 site is indicated on the x axis. P values are as follows: Q555X versus untreated, P = 0.001001 (double asterisk); E4 splice versus untreated, P = 0.001295 (double asterisk). c, Relative change in LDL-R present in supernatant between day 9 and day 0 assessed by ELISA. P values are as follows: Q555X versus untreated, P = 0.001116 (double asterisk), E4 splice versus untreated, P = .009481 (double asterisk). d, Gene-editing efficiencies from sgRNA screens in primary human T cells using CBE-Ts. X axis label indicates the targeted gene and target base within sgRNA. e,f, Percent C·G to T·A conversion (e) and surface protein loss (f) achieved by each base editor or control in multiplex-edited primary human T cells. Primary hepatocyte data were generated from n = 3 (‘none’/untreated samples) or 4 (BE4 and CBE-T1.46 samples) independent biological replicates. T cell data were generated from n = 2 independent donors. Where applicable, statistical significance was computed via two-tailed unpaired t tests: NS, P ≥ 0.05; *P < 0.05; **P < 0.01.