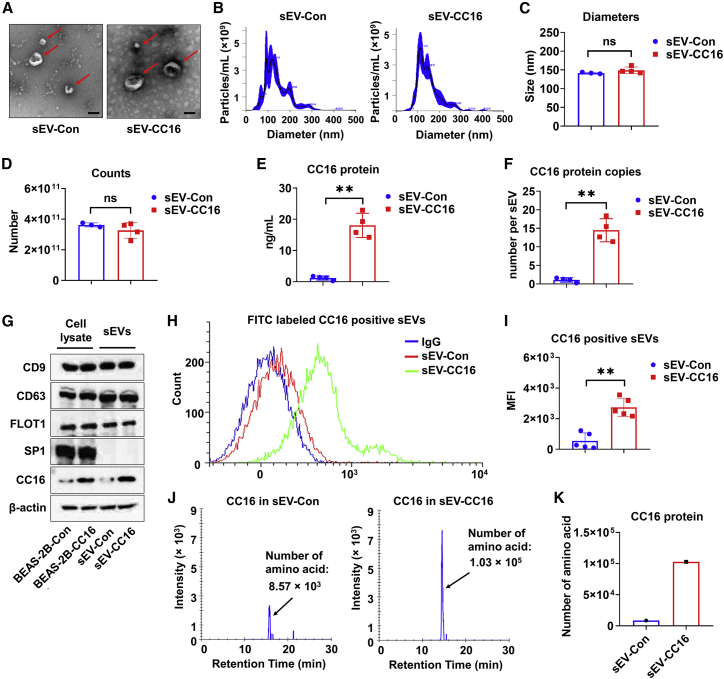
Figure 2.
Preparation and characterization of sEV-Con and sEV-CC16
(A) TEM images of sEV-Con and sEV-CC16 isolated from culture media of BEAS-2B-Con and BEAS-2B-CC16. Scale bar, 100 μm. (B–D) Two-hundred micrograms of sEV-Con and sEV-CC16 are characterized using NTA for size distribution (B), diameter (C), and particle number (D). Results are represented as mean ± SD of a minimum of three independent experiments. ns, p > 0.05; ∗∗p < 0.01. (E) The CC16 protein amount in 200 μg of sEV-Con and sEV-CC16 is quantified using ELISA (n = 4). (F) Copy number of CC16 in single sEV-Con and sEV-CC16 is calculated (n = 4). Results are presented as mean ± SD. ∗∗p < 0.01. (G) sEV positive markers (CD9, CD63, and Flot1) and a negative marker (Sp1) are detected in 50 μg of protein from BEAS-2B-Con, BEAS-2B-CC16, sEV-Con, and sEV-CC16 using western blot (n = 3). (H and I) Detection of CC16-positive sEVs from sEV-Con and sEV-CC16 using flow cytometry (n = 5). Results are presented as mean ± SD. ∗∗p < 0.01. (J and K) sEV-Con and sEV-CC16 isolated from five independent sEV isolation processes were pooled and analyzed by MS. The number of CC16-derived amino acids in 20 μg of pooled sEV-Con and sEV-CC16 are quantified.