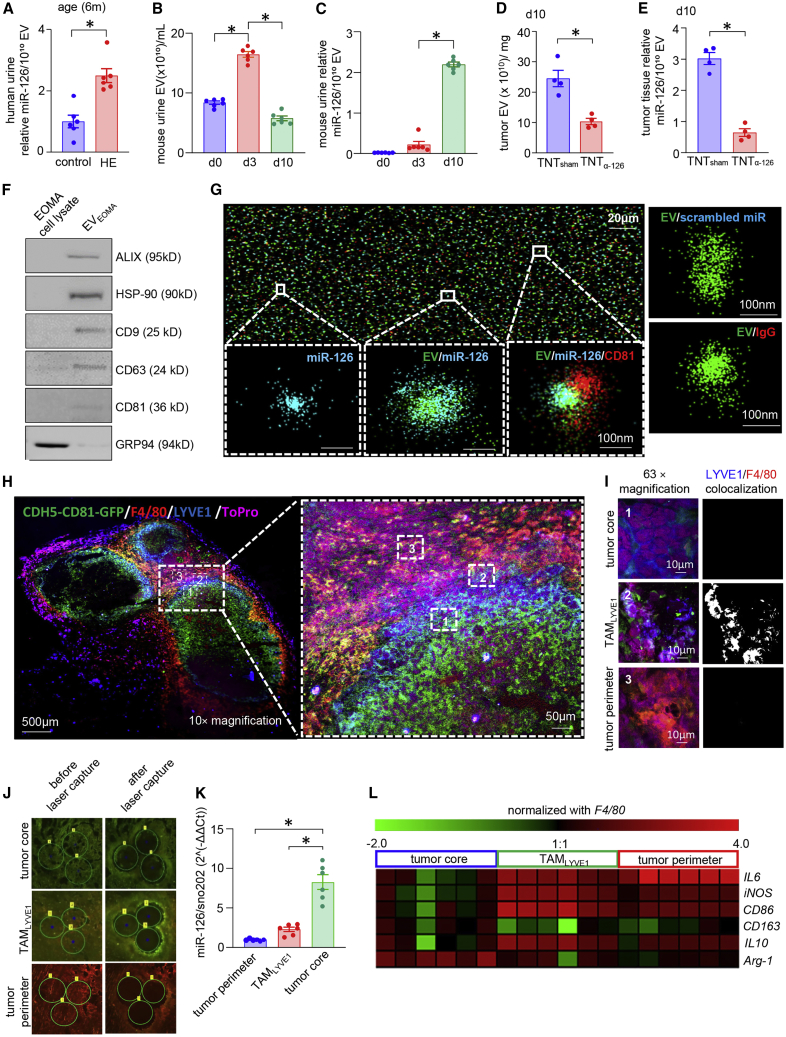
Figure 2.
Characterization of EOMA-derived EVs and their preferential uptake by TAMs
(A) miR-126 expression in the extracellular vesicles (EVs) isolated from urine samples from age-matched healthy and HE patients (6 months old, n = 6), ∗p < 0.05, Student’s t test. (B) Abundance of EVs from mouse urine isolated from different days after tumor progression, n = 6, ∗p < 0.05, one-way ANOVA. (C) miR-126 abundance in EVs isolated from murine urine samples in different days of tumor development. ∗p < 0.05 n = 6, one-way ANOVA. (D) Quantification of EVs, estimated from tumor tissue (d10) after treatment with scrambled oligonucleotides (TNTsham) or antagomir-126 (TNTα-miR-126). ∗p < 0.05 (n = 4), Mann Whitney test. (E) Abundance of miR-126 from the isolated EV samples collected from tissue after TNTα-miR-126 or TNTsham treatment. ∗p < 0.05, n = 4, Mann Whitney test. (F) Western blot analysis of exosome markers Alix, HSP90, CD9, CD63, CD81, and exosome-negative marker GRP94. (G) Super-resolution microscopic image shows Cy3-labeled miR-126 mimic cargo present inside Atto 488-labeled exosome isolated from EOMA cell culture media. Red represents Alexa 647-tagged CD81. Eighty-two percent fraction of EVEOMA containing miR-126 cargo. Cy3 scrambled control oligonucleotide for miRNA and Alexa Fluor 647-labeled IgG antibody-stained EOMA EVs were used as negative controls. (H) Injection of CDH5-CD81-GFP-transduced EOMA cells into mice formed HE tumors in which EVs secreted by EOMA cells expressed GFP. Immunohistochemical analysis showing co-expression of F4/80 (Mφ marker, red), LYVE1 (EOMA cell marker, blue) with GFP expressed by EVEOMA (green). Nucleus is counter-stained with ToPro3. Representative confocal image is taken at 10× magnification (left) and its expansion (right) representing different regions of tumor, (i) MφEOMA-deficient tumor core region (tumor core), (ii) intermediary LYVE1+ TAM-populated region (TAMLYVE1), and (iii) MφEOMA-deficient tumor perimeter region (tumor perimeter). (I) Panel represents magnification (63×) of these three regions and extent of colocalization between F4/80 and LYVE1. Single-channel images of F4/80/LYVE1 colocalization is provided in Figure S4C. (J) Laser capture microdissection (LCM) images from different regions of tumor tissue sections before and after cut. (K) Abundance of miR-126 in different regions of HE tumor captured using LCM. ∗p < 0.05, n = 6, one-way ANOVA. (L) Heatmap representing the expression of M1 and M2 macrophage markers in different HE tumor regions captured through LCM. Data presented as mean ± SEM.