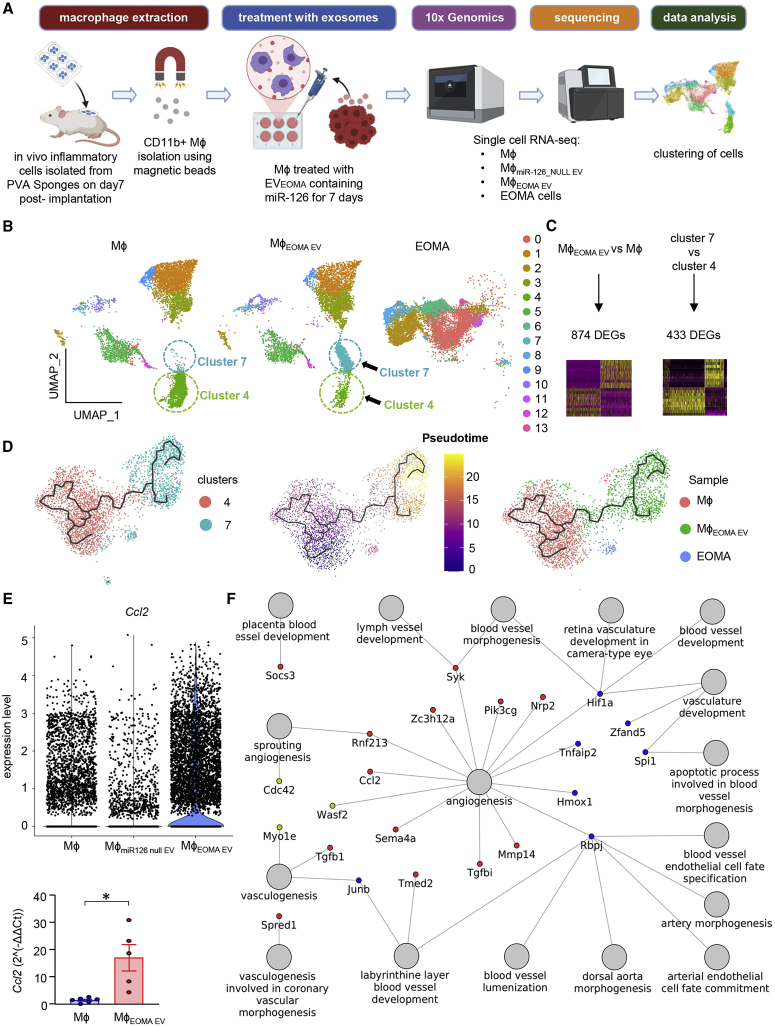
Figure 4.
Single-cell RNA sequencing data analysis of LYVE+ tumorigenic TAMs cells identified the cellular trajectories responsible for tumor growth
(A) Schematic diagram of sample collection and single-cell analysis workflow. (B) Uniform Manifold Approximation and Projection (UMAP) of the filtered data (23,250 cells) clustered into 14 clusters. Each cell is represented as a dot. (C) Schematic representation of the differential expression analysis performed between all EVEOMA treated versus all untreated Mφ and between cluster 7 versus cluster 4. (D) Trajectory inference of cells from clusters 4 and 7 colored by clusters (left), pseudotime (middle), or the originating sample (right). (E) Violin plots representing expression level of Ccl2 in Mφ, MφmiR126 null EV, and in MφEOMA EV cells (upper). Bar plots of PCR results for Ccl2 (below). (F) Network of upregulated genes found in both comparisons Mφ EOMA EV versus Mφ or in cluster 7 versus cluster 4 cells. Nodes with red colors represent genes found to be upregulated in both comparisons MφEOMA EV versus mφ and cluster 7 cells versus cluster 4 cells. Nodes with blue color represent genes found to be upregulated in MφEOMA EV versus Mφ cells, but not in cluster 7 cells versus cluster 4 cells. Nodes with golden color represent genes found to be upregulated in cluster 7 cells versus cluster 4 cells, but not in MφEOMA EV versus Mφ cells. Data presented as mean ± SEM.