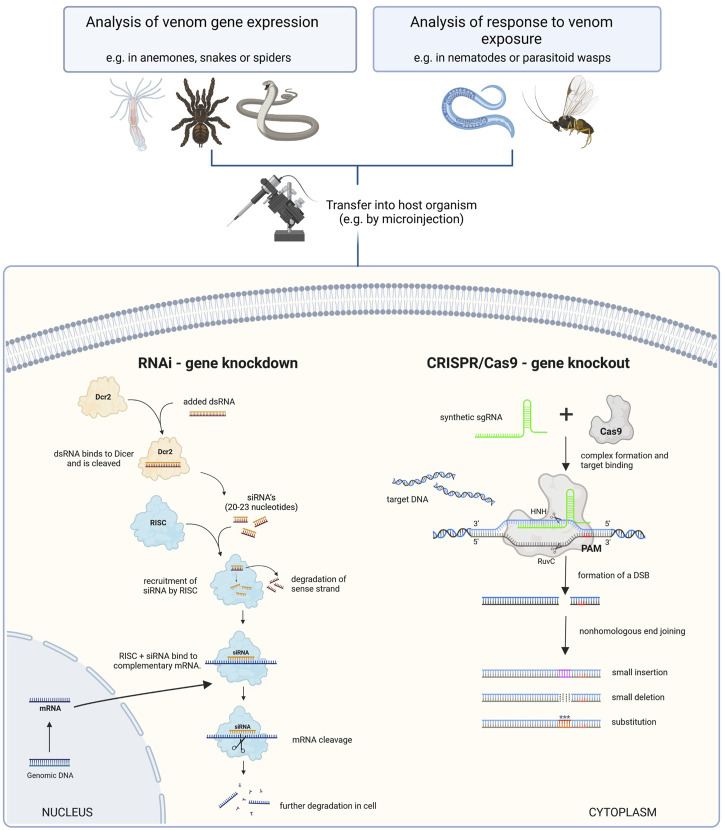
FIGURE 2.
Comparison of functional genomics tools that enable loss-of-function studies in venom research. Left: RNAi is used to induce the temporary knockdown of targeted genes by degrading mRNAs. At first double-stranded RNA (dsRNA) is injected and binds to the Dicer protein, where it is cleaved into small interfering RNAs (siRNA). These are subsequentially recruited to the RNA-induced silencing complex (RISC), which binds to complementary mRNA and facilitates its degradation. Thus, the corresponding gene is temporarily not translated. Right: CRISPR/Cas9 is typically used to introduce double-strand breaks that are erroneously repaired, leading to a stable knockout mutation. Here, single guide RNA (sgRNA) is injected and binds to the Cas9 protein. The resulting complex interacts with target DNA complementary to the sgRNA. Cas9 cleaves the DNA upstream to the protospacer adjacent motif (PAM) and leads to nonhomologous end joining, allowing to precisely add substitutions, smaller insertions and deletions to the sequence in dependence of experimental conditions. Deletions lead to permanent removal of targeted sequences and thus the corresponding gene can not be expressed. The figure is adapted from (Knorr et al., 2013; Jiang and Doudna, 2017; Bharathkumar et al., 2021).