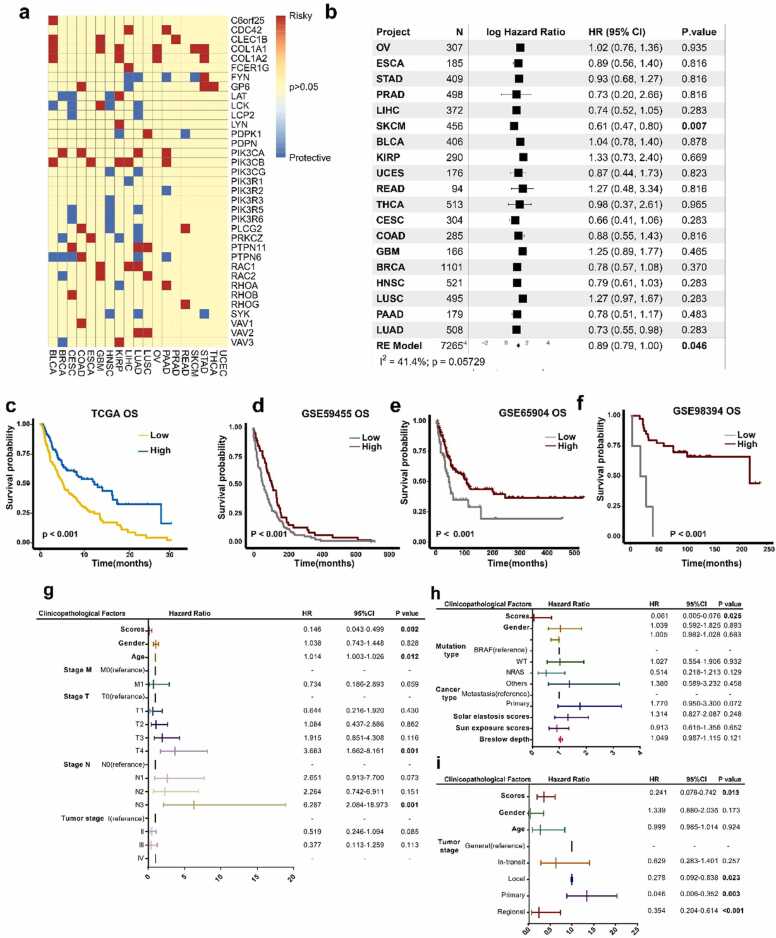
Fig. 2.
Identification of GMPA signatures associated with patient survival. (a) Prognostic value of GMPA signature-related genes for OS in 19 types of cancer in TCGA. (b) (b) Forest plots depicting results of Cox proportional hazards regression for OS analysis using GMPA signature scores. A random effects model was used to calculate the pooled hazard ratio and the p-values. Forest plots showing loge hazard ratio (95% confidence interval). P-values were adjusted for multiple testing using the FDR method (significance threshold of 5%). I-square (I2) statistic test was used to evaluate the proportion of statistical heterogeneity. (c-f) K-M curves of OS by GMPA signature scores in TCGA (c), GSE59455 (d), GSE65904 (e), and GSE98394 (f) datasets. P values for survival analysis were calculated using log rank tests. (g-i) Multivariate regression analysis of OS in patients with SKCM from TCGA (g), GSE65904 (h), and GSE59455 datasets (i). P-values less than 0.05 are in bold in the Figure.