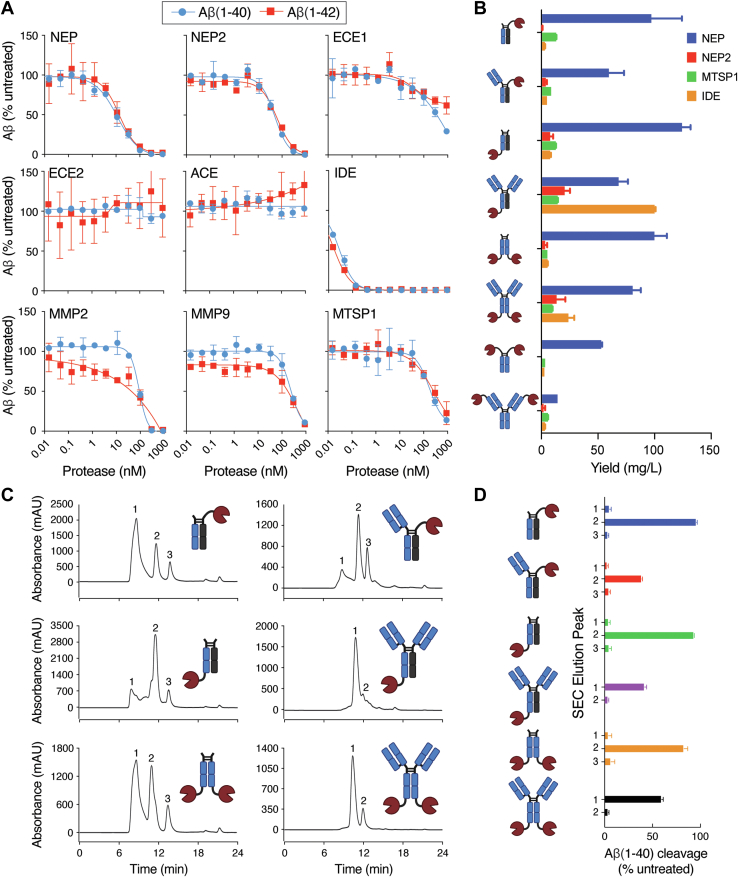
Figure 2.
Protease and fusion format screening for targeted degradation of Aβ. A, in vitro protease screening assay for cleavage of Aβ(1–40) (blue circles) and Aβ(1–42) (red squares). B, expression yields of Fc and IgG protease fusion formats. Four different proteases were expressed in the context of the eight formats shown in the icons. IgG fusions contained variable regions of the anti-Aβ antibody crenezumab, and all heavy chain constant regions were human IgG1. The bar graph shows the expression yields from duplicate 30 ml HEK293 expressions of each construct. C, purification of crenezumab NEP protease fusion formats. Each NEP fusion format was expressed in HEK293 cells and initially purified using a protein A resin. Size exclusion chromatography (SEC) coupled with sample fractionation was used for further purification. SEC chromatograms revealed the presence of multiple species with each sample containing 2 to 3 peaks. Nonreducing SDS-PAGE analysis to identify the peaks of similar antibody–protease fusions is shown in Fig. S7. D, the central fraction associated with each peak in the chromatograms from (C) was tested for Aβ(1–40) cleavage, and fractions of active peaks were pooled to obtain samples free of unwanted species. Error bars in (A, B, and D) represent standard error values with n = 2. Aβ, amyloid-β; ECE, endothelin-converting enzyme; Fc, fragment crystallizable; IgG, immunoglobulin G; IDE, insulin-degrading enzyme; MMP, matrix metalloproteinase; MTSP1, matriptase; NEP, neprolysin.